Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for HOXB5
Z-value: 0.46
Transcription factors associated with HOXB5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB5
|
ENSG00000120075.6 | HOXB5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB5 | hg38_v1_chr17_-_48593748_48593786 | 0.21 | 1.4e-03 | Click! |
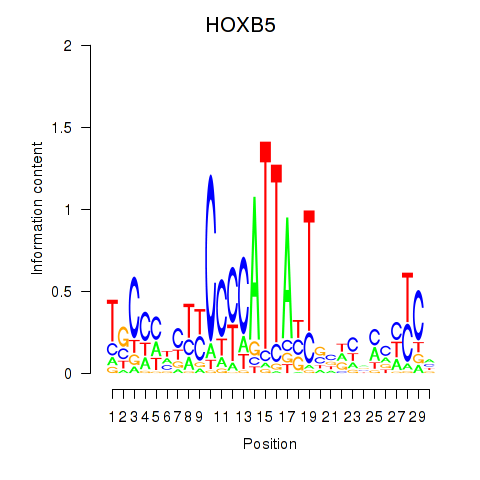
Activity profile of HOXB5 motif
Sorted Z-values of HOXB5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_48521817 | 11.73 |
ENST00000446158.5
ENST00000414061.1 |
EBP
|
EBP cholestenol delta-isomerase |
| chrX_+_48521788 | 7.71 |
ENST00000651615.1
ENST00000495186.6 |
ENSG00000286268.1
EBP
|
novel protein EBP cholestenol delta-isomerase |
| chr19_-_49640092 | 6.22 |
ENST00000246792.4
|
RRAS
|
RAS related |
| chr14_-_102238439 | 5.40 |
ENST00000522534.5
|
MOK
|
MOK protein kinase |
| chr15_+_73926443 | 4.40 |
ENST00000261921.8
|
LOXL1
|
lysyl oxidase like 1 |
| chr2_-_46941710 | 3.97 |
ENST00000409207.5
|
MCFD2
|
multiple coagulation factor deficiency 2, ER cargo receptor complex subunit |
| chr1_-_161223408 | 3.16 |
ENST00000491350.1
|
APOA2
|
apolipoprotein A2 |
| chr1_-_161223559 | 3.15 |
ENST00000469730.2
ENST00000463273.5 ENST00000464492.5 ENST00000367990.7 ENST00000470459.6 ENST00000463812.1 ENST00000468465.5 |
APOA2
|
apolipoprotein A2 |
| chr3_-_127822455 | 2.64 |
ENST00000265052.10
|
MGLL
|
monoglyceride lipase |
| chr3_+_51385282 | 2.29 |
ENST00000528157.7
|
MANF
|
mesencephalic astrocyte derived neurotrophic factor |
| chr2_-_46941760 | 2.27 |
ENST00000444761.6
ENST00000409147.1 |
MCFD2
|
multiple coagulation factor deficiency 2, ER cargo receptor complex subunit |
| chr19_-_15934410 | 2.14 |
ENST00000326742.12
|
CYP4F11
|
cytochrome P450 family 4 subfamily F member 11 |
| chr8_-_132085655 | 2.11 |
ENST00000262283.5
|
ENSG00000258417.3
|
novel protein |
| chr12_+_53299682 | 2.09 |
ENST00000267103.10
ENST00000548632.5 |
MYG1
|
MYG1 exonuclease |
| chr1_-_16978276 | 2.01 |
ENST00000375534.7
|
MFAP2
|
microfibril associated protein 2 |
| chr1_+_21981099 | 1.74 |
ENST00000400277.2
|
CELA3B
|
chymotrypsin like elastase 3B |
| chr7_+_130381092 | 1.69 |
ENST00000484324.1
|
CPA1
|
carboxypeptidase A1 |
| chr1_+_154257071 | 1.68 |
ENST00000428595.1
|
UBAP2L
|
ubiquitin associated protein 2 like |
| chr16_-_50368920 | 1.67 |
ENST00000394688.8
|
BRD7
|
bromodomain containing 7 |
| chrX_+_47218232 | 1.64 |
ENST00000457458.6
ENST00000522883.1 |
CDK16
|
cyclin dependent kinase 16 |
| chr16_-_84618067 | 1.53 |
ENST00000262428.5
|
COTL1
|
coactosin like F-actin binding protein 1 |
| chr15_-_74725370 | 1.52 |
ENST00000567032.5
ENST00000564596.5 ENST00000566503.1 ENST00000395049.8 ENST00000379727.8 ENST00000617691.4 ENST00000395048.6 |
CYP1A1
|
cytochrome P450 family 1 subfamily A member 1 |
| chr1_+_40691749 | 1.51 |
ENST00000372654.5
|
NFYC
|
nuclear transcription factor Y subunit gamma |
| chr20_-_49568101 | 1.49 |
ENST00000244043.5
|
PTGIS
|
prostaglandin I2 synthase |
| chr1_+_155610218 | 1.41 |
ENST00000649846.1
ENST00000245564.8 ENST00000368341.8 |
MSTO1
|
misato mitochondrial distribution and morphology regulator 1 |
| chr1_+_40691998 | 1.38 |
ENST00000534399.5
ENST00000372653.5 |
NFYC
|
nuclear transcription factor Y subunit gamma |
| chr12_+_57591158 | 1.38 |
ENST00000422156.7
ENST00000354947.10 ENST00000540759.6 ENST00000551772.5 ENST00000550465.5 |
PIP4K2C
|
phosphatidylinositol-5-phosphate 4-kinase type 2 gamma |
| chr1_-_155688294 | 1.37 |
ENST00000311573.9
|
YY1AP1
|
YY1 associated protein 1 |
| chr19_-_15934521 | 1.35 |
ENST00000402119.9
|
CYP4F11
|
cytochrome P450 family 4 subfamily F member 11 |
| chr9_-_127937800 | 1.34 |
ENST00000373110.4
ENST00000314392.13 |
DPM2
|
dolichyl-phosphate mannosyltransferase subunit 2, regulatory |
| chr16_-_84618041 | 1.32 |
ENST00000564057.1
|
COTL1
|
coactosin like F-actin binding protein 1 |
| chr17_-_44066595 | 1.20 |
ENST00000585388.2
ENST00000293406.8 |
LSM12
|
LSM12 homolog |
| chr16_-_1771495 | 1.12 |
ENST00000564628.1
ENST00000219302.8 ENST00000563498.1 |
NME3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr17_+_48892761 | 1.11 |
ENST00000355938.9
ENST00000393366.7 ENST00000503641.5 ENST00000514808.5 ENST00000506855.1 |
ATP5MC1
|
ATP synthase membrane subunit c locus 1 |
| chr1_+_235328959 | 1.09 |
ENST00000643758.1
ENST00000497327.1 |
TBCE
GGPS1
|
tubulin folding cofactor E geranylgeranyl diphosphate synthase 1 |
| chr12_+_56521798 | 0.97 |
ENST00000262031.10
|
RBMS2
|
RNA binding motif single stranded interacting protein 2 |
| chr17_-_30906202 | 0.91 |
ENST00000580840.1
ENST00000581216.6 |
TEFM
|
transcription elongation factor, mitochondrial |
| chr1_-_155688475 | 0.90 |
ENST00000405763.7
ENST00000368340.9 ENST00000454523.5 ENST00000368339.9 ENST00000443231.5 ENST00000347088.9 ENST00000361831.9 |
YY1AP1
|
YY1 associated protein 1 |
| chr20_+_8789517 | 0.85 |
ENST00000437439.2
|
PLCB1
|
phospholipase C beta 1 |
| chr12_+_56521990 | 0.79 |
ENST00000550726.5
ENST00000542360.1 |
RBMS2
|
RNA binding motif single stranded interacting protein 2 |
| chr6_-_31541937 | 0.77 |
ENST00000456662.5
ENST00000431908.5 ENST00000456976.5 ENST00000428450.5 ENST00000418897.5 ENST00000396172.6 ENST00000419020.1 ENST00000428098.5 |
DDX39B
|
DExD-box helicase 39B |
| chr12_+_56521951 | 0.75 |
ENST00000552247.6
|
RBMS2
|
RNA binding motif single stranded interacting protein 2 |
| chr3_-_195811889 | 0.75 |
ENST00000475231.5
|
MUC4
|
mucin 4, cell surface associated |
| chr19_+_49363730 | 0.72 |
ENST00000596402.1
ENST00000221498.7 |
DKKL1
|
dickkopf like acrosomal protein 1 |
| chr3_-_195811916 | 0.72 |
ENST00000463781.8
|
MUC4
|
mucin 4, cell surface associated |
| chr2_-_97589862 | 0.71 |
ENST00000258459.11
ENST00000438709.6 |
ANKRD36B
|
ankyrin repeat domain 36B |
| chr12_-_101407727 | 0.69 |
ENST00000539055.5
ENST00000551688.1 ENST00000551671.5 ENST00000261636.13 |
ARL1
|
ADP ribosylation factor like GTPase 1 |
| chr1_+_40691689 | 0.66 |
ENST00000427410.6
ENST00000447388.7 ENST00000425457.6 ENST00000453631.5 ENST00000456393.6 |
NFYC
|
nuclear transcription factor Y subunit gamma |
| chr3_-_195811857 | 0.65 |
ENST00000349607.8
ENST00000346145.8 |
MUC4
|
mucin 4, cell surface associated |
| chr1_-_155688727 | 0.62 |
ENST00000404643.5
ENST00000359205.9 ENST00000407221.5 ENST00000355499.9 |
YY1AP1
|
YY1 associated protein 1 |
| chr6_-_31542339 | 0.59 |
ENST00000458640.5
|
DDX39B
|
DExD-box helicase 39B |
| chr6_-_96837460 | 0.57 |
ENST00000229955.4
|
GPR63
|
G protein-coupled receptor 63 |
| chr21_-_44262216 | 0.57 |
ENST00000270172.7
|
DNMT3L
|
DNA methyltransferase 3 like |
| chr2_-_187565751 | 0.57 |
ENST00000421427.5
|
TFPI
|
tissue factor pathway inhibitor |
| chr1_+_4654732 | 0.55 |
ENST00000378190.7
|
AJAP1
|
adherens junctions associated protein 1 |
| chr19_-_49763295 | 0.53 |
ENST00000246801.8
|
TSKS
|
testis specific serine kinase substrate |
| chr1_+_235328399 | 0.50 |
ENST00000471812.5
ENST00000358966.6 ENST00000282841.9 ENST00000391855.2 ENST00000647428.1 ENST00000645655.1 ENST00000647186.1 ENST00000646624.1 ENST00000645205.1 ENST00000645351.1 |
GGPS1
TBCE
|
geranylgeranyl diphosphate synthase 1 tubulin folding cofactor E |
| chrX_-_107982370 | 0.49 |
ENST00000302917.1
|
TEX13B
|
testis expressed 13B |
| chr19_+_49363923 | 0.48 |
ENST00000597546.1
|
DKKL1
|
dickkopf like acrosomal protein 1 |
| chr1_+_24502320 | 0.44 |
ENST00000538532.6
ENST00000618490.4 |
RCAN3
|
RCAN family member 3 |
| chr1_+_116373233 | 0.36 |
ENST00000295598.10
|
ATP1A1
|
ATPase Na+/K+ transporting subunit alpha 1 |
| chrM_+_12329 | 0.32 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 5 |
| chrX_+_14529516 | 0.30 |
ENST00000218075.9
|
GLRA2
|
glycine receptor alpha 2 |
| chr10_-_101843765 | 0.28 |
ENST00000370046.5
|
KCNIP2
|
potassium voltage-gated channel interacting protein 2 |
| chr8_-_92095598 | 0.28 |
ENST00000520724.5
ENST00000518844.5 |
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr1_+_161153968 | 0.27 |
ENST00000368003.6
|
UFC1
|
ubiquitin-fold modifier conjugating enzyme 1 |
| chr14_+_21477177 | 0.26 |
ENST00000448790.7
ENST00000673643.1 ENST00000457430.2 ENST00000673911.1 |
TOX4
|
TOX high mobility group box family member 4 |
| chr11_-_35419098 | 0.25 |
ENST00000606205.6
ENST00000645303.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr12_-_7665897 | 0.19 |
ENST00000229304.5
|
APOBEC1
|
apolipoprotein B mRNA editing enzyme catalytic subunit 1 |
| chr1_+_4654601 | 0.18 |
ENST00000378191.5
|
AJAP1
|
adherens junctions associated protein 1 |
| chr6_+_29459771 | 0.18 |
ENST00000377132.1
|
OR2H1
|
olfactory receptor family 2 subfamily H member 1 |
| chr1_+_157993601 | 0.17 |
ENST00000359209.11
|
KIRREL1
|
kirre like nephrin family adhesion molecule 1 |
| chr11_-_128522189 | 0.15 |
ENST00000526145.6
|
ETS1
|
ETS proto-oncogene 1, transcription factor |
| chr19_-_13953302 | 0.09 |
ENST00000585607.1
ENST00000538517.6 ENST00000587458.1 ENST00000538371.6 |
PODNL1
|
podocan like 1 |
| chr10_+_112376193 | 0.09 |
ENST00000433418.6
ENST00000356116.6 ENST00000354273.5 |
ACSL5
|
acyl-CoA synthetase long chain family member 5 |
| chr7_+_121328991 | 0.09 |
ENST00000222462.3
|
WNT16
|
Wnt family member 16 |
| chr17_-_7207245 | 0.08 |
ENST00000649971.1
|
DLG4
|
discs large MAGUK scaffold protein 4 |
| chr16_+_31527876 | 0.07 |
ENST00000302312.9
ENST00000564707.1 |
AHSP
|
alpha hemoglobin stabilizing protein |
| chr4_-_67754335 | 0.06 |
ENST00000420975.2
ENST00000226413.5 |
GNRHR
|
gonadotropin releasing hormone receptor |
| chr8_-_92095215 | 0.05 |
ENST00000360348.6
ENST00000520428.5 ENST00000518992.5 ENST00000520556.5 ENST00000518317.5 ENST00000521319.5 ENST00000521375.5 ENST00000518449.5 ENST00000613886.4 |
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chrX_+_136148440 | 0.03 |
ENST00000627383.2
ENST00000630084.2 |
FHL1
|
four and a half LIM domains 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 19.4 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 1.6 | 6.3 | GO:0060620 | regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 0.9 | 3.5 | GO:0042361 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.5 | 1.6 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.5 | 1.5 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 0.3 | 2.6 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.3 | 2.0 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.3 | 0.8 | GO:0007343 | egg activation(GO:0007343) |
| 0.3 | 1.3 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.2 | 1.7 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.2 | 1.4 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 6.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 1.4 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.1 | 1.5 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.1 | 0.7 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.1 | 6.2 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 2.1 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.9 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.7 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 2.9 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.1 | 1.4 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.1 | 0.6 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.6 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 4.4 | GO:0035904 | aorta development(GO:0035904) |
| 0.1 | 2.3 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 1.1 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 1.6 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 3.6 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 0.4 | GO:1903416 | negative regulation of glucocorticoid metabolic process(GO:0031944) negative regulation of glucocorticoid biosynthetic process(GO:0031947) negative regulation of steroid hormone biosynthetic process(GO:0090032) response to glycoside(GO:1903416) |
| 0.0 | 0.3 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 1.7 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.3 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.2 | GO:0070383 | cytidine to uridine editing(GO:0016554) DNA cytosine deamination(GO:0070383) regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.0 | 0.1 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) |
| 0.0 | 0.1 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 1.2 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.2 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.0 | 0.2 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 1.7 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.4 | 6.3 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.3 | 3.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 2.9 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 0.7 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 5.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 1.4 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 2.0 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 1.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 6.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 4.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.7 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 20.3 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 0.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.9 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 2.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.5 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 2.9 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 4.4 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 6.3 | GO:0005925 | focal adhesion(GO:0005925) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.5 | 19.4 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 1.3 | 6.3 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.5 | 1.4 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.4 | 1.3 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.4 | 2.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.3 | 1.6 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.3 | 1.5 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.3 | 0.9 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.2 | 4.4 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.2 | 2.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 1.4 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.1 | 7.0 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 6.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 3.5 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 0.3 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 1.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 1.7 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 1.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 3.6 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.4 | GO:1990239 | sodium:potassium-exchanging ATPase activity(GO:0005391) steroid hormone binding(GO:1990239) |
| 0.0 | 1.5 | GO:0016860 | intramolecular oxidoreductase activity(GO:0016860) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 19.4 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 5.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.8 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 3.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 2.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 5.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 21.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.3 | 6.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.3 | 6.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 6.2 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 6.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 2.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 0.8 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.6 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |