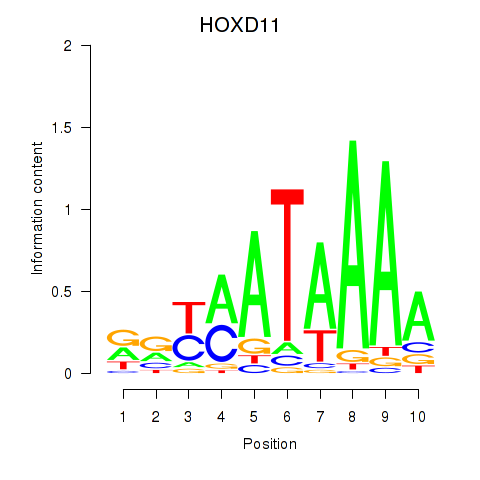
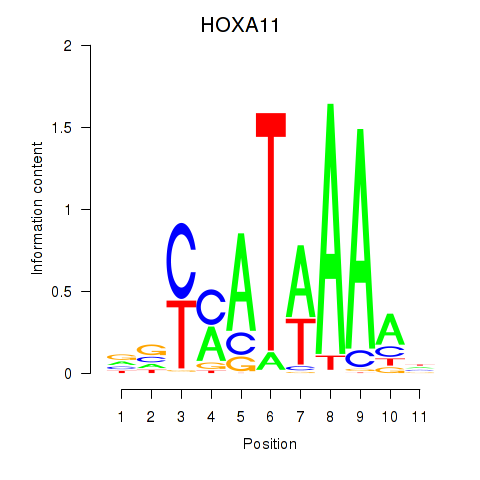
Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for HOXD11_HOXA11
Z-value: 0.76
Transcription factors associated with HOXD11_HOXA11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD11
|
ENSG00000128713.14 | HOXD11 |
|
HOXA11
|
ENSG00000005073.6 | HOXA11 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA11 | hg38_v1_chr7_-_27185223_27185249 | -0.28 | 2.9e-05 | Click! |
| HOXD11 | hg38_v1_chr2_+_176107272_176107297 | -0.19 | 4.4e-03 | Click! |
Activity profile of HOXD11_HOXA11 motif
Sorted Z-values of HOXD11_HOXA11 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD11_HOXA11
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 9.1 | GO:0070476 | RNA (guanine-N7)-methylation(GO:0036265) rRNA (guanine-N7)-methylation(GO:0070476) |
| 2.7 | 11.0 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 2.7 | 8.1 | GO:1902594 | viral penetration into host nucleus(GO:0075732) multi-organism nuclear import(GO:1902594) |
| 1.7 | 26.0 | GO:0000338 | protein deneddylation(GO:0000338) |
| 1.6 | 4.7 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 1.6 | 9.3 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 1.4 | 6.9 | GO:2000542 | negative regulation of gastrulation(GO:2000542) |
| 1.4 | 16.4 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 1.3 | 5.1 | GO:0036114 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 1.2 | 3.6 | GO:1901993 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 1.2 | 3.5 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 1.1 | 19.3 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 1.1 | 4.5 | GO:0050923 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 1.1 | 5.5 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 1.1 | 5.3 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 1.1 | 5.3 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 1.0 | 4.2 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.9 | 2.8 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.9 | 3.7 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.9 | 5.4 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.8 | 16.8 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.8 | 4.6 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.8 | 3.8 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.8 | 2.3 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.7 | 2.9 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.7 | 4.3 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.7 | 3.5 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.7 | 9.2 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.7 | 2.1 | GO:0051793 | medium-chain fatty acid catabolic process(GO:0051793) |
| 0.7 | 4.8 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.7 | 2.0 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.7 | 4.0 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.7 | 3.3 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.7 | 2.0 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.6 | 2.6 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.6 | 3.2 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.6 | 1.9 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.6 | 2.5 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.6 | 1.8 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.6 | 3.5 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.6 | 4.5 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.6 | 3.3 | GO:1903936 | cellular response to sodium arsenite(GO:1903936) |
| 0.6 | 3.3 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.5 | 2.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.5 | 3.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.5 | 3.0 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.5 | 3.0 | GO:0006548 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.5 | 1.5 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.5 | 2.4 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.5 | 2.3 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.5 | 2.3 | GO:0045338 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) farnesyl diphosphate metabolic process(GO:0045338) |
| 0.5 | 1.4 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.4 | 1.3 | GO:1901388 | regulation of transforming growth factor beta activation(GO:1901388) negative regulation of transforming growth factor beta activation(GO:1901389) |
| 0.4 | 3.5 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.4 | 2.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.4 | 13.5 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.4 | 4.5 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.4 | 5.7 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.4 | 23.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.4 | 1.6 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of G-protein coupled receptor internalization(GO:1904020) negative regulation of G-protein coupled receptor internalization(GO:1904021) |
| 0.4 | 4.7 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.4 | 3.1 | GO:0046459 | short-chain fatty acid metabolic process(GO:0046459) |
| 0.4 | 4.1 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.4 | 1.5 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.4 | 1.8 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.3 | 1.6 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.3 | 4.7 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.3 | 1.9 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.3 | 6.0 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.3 | 9.6 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.3 | 2.2 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.3 | 3.5 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.3 | 1.9 | GO:0032328 | alanine transport(GO:0032328) |
| 0.3 | 22.8 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.3 | 2.6 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.3 | 1.6 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.3 | 1.5 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.2 | 3.2 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.2 | 6.1 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.2 | 2.4 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.2 | 1.7 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.2 | 1.9 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.2 | 2.3 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.2 | 3.5 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.2 | 1.8 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.2 | 4.4 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.2 | 4.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.2 | 1.9 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.2 | 2.6 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.2 | 1.1 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.2 | 0.4 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.2 | 1.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.2 | 4.6 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.2 | 2.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.2 | 1.2 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.2 | 5.5 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.2 | 3.7 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.2 | 0.9 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.2 | 8.2 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.2 | 1.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.2 | 0.7 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.2 | 1.5 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.2 | 2.2 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.2 | 0.5 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.2 | 1.8 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.2 | 2.6 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.2 | 2.2 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.2 | 1.0 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 | 1.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.2 | 1.2 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.2 | 0.8 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.2 | 0.9 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.1 | 0.7 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.1 | 3.8 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 1.6 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 | 2.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 2.0 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.1 | 2.6 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) |
| 0.1 | 2.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 8.0 | GO:0032873 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.1 | 0.5 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.1 | 5.7 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.1 | 1.1 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.1 | 1.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.4 | GO:0018283 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.1 | 2.5 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.1 | 3.7 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 1.4 | GO:0070989 | oxidative demethylation(GO:0070989) |
| 0.1 | 1.6 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 1.4 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 2.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 20.2 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.1 | 8.1 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.1 | 9.7 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.1 | 0.8 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 0.5 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.1 | 0.3 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.1 | 2.4 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 0.1 | 1.2 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 1.0 | GO:0032930 | positive regulation of superoxide anion generation(GO:0032930) |
| 0.1 | 1.2 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.1 | 3.3 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 3.7 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 2.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 1.0 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.4 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.3 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 0.3 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.1 | 0.3 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 0.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 14.6 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 | 0.4 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.1 | 0.5 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 2.8 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 8.0 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.2 | GO:0060003 | copper ion export(GO:0060003) |
| 0.1 | 0.3 | GO:0010710 | regulation of collagen catabolic process(GO:0010710) |
| 0.1 | 0.5 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 1.5 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.8 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.1 | 1.9 | GO:0007099 | centriole replication(GO:0007099) centriole assembly(GO:0098534) |
| 0.1 | 2.7 | GO:0007031 | peroxisome organization(GO:0007031) |
| 0.1 | 0.5 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.9 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.7 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.5 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 1.2 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.9 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 3.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 3.5 | GO:0009154 | purine ribonucleotide catabolic process(GO:0009154) |
| 0.0 | 0.3 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.3 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.4 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.0 | 0.6 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 6.6 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 0.8 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.3 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 1.5 | GO:0030516 | regulation of axon extension(GO:0030516) |
| 0.0 | 0.4 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 1.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.7 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.0 | 0.6 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 1.0 | GO:0032986 | nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 1.6 | GO:0032480 | negative regulation of type I interferon production(GO:0032480) |
| 0.0 | 0.9 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.0 | 0.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.5 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.9 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 2.0 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 1.9 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 4.6 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.4 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.9 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.4 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.7 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.4 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 | 0.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.6 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 5.3 | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 1.0 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 1.1 | GO:0031640 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.0 | 2.1 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 1.1 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.3 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 2.4 | GO:0030307 | positive regulation of cell growth(GO:0030307) |
| 0.0 | 1.0 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.3 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.4 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.6 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.4 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.3 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.2 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 23.4 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 4.4 | 17.7 | GO:1990423 | RZZ complex(GO:1990423) |
| 2.3 | 6.9 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.7 | 17.3 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.9 | 5.7 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.9 | 5.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.8 | 8.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.8 | 5.5 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.8 | 2.3 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.7 | 2.0 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.7 | 2.0 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.6 | 1.9 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.6 | 11.9 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.6 | 30.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.5 | 2.6 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.5 | 5.8 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.5 | 1.9 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.5 | 9.8 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.4 | 2.2 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.4 | 9.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.4 | 6.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.4 | 8.0 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.4 | 3.0 | GO:0000796 | condensin complex(GO:0000796) |
| 0.4 | 11.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.4 | 1.8 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.3 | 6.4 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.3 | 1.6 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.3 | 6.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.3 | 5.0 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.3 | 1.6 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.3 | 1.8 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.2 | 1.5 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.2 | 16.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.2 | 0.9 | GO:0035363 | histone locus body(GO:0035363) |
| 0.2 | 10.8 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.2 | 1.9 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.2 | 4.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.2 | 1.2 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.2 | 2.8 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.2 | 4.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 1.6 | GO:0070522 | nucleotide-excision repair factor 1 complex(GO:0000110) ERCC4-ERCC1 complex(GO:0070522) |
| 0.2 | 2.1 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 1.8 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.2 | 2.9 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.2 | 2.8 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.2 | 0.5 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.2 | 2.9 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 2.6 | GO:0034709 | methylosome(GO:0034709) |
| 0.2 | 1.8 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.2 | 0.9 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.2 | 1.5 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.2 | 1.7 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 1.9 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 6.0 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 2.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 2.7 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 7.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 4.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 2.8 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 1.2 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 14.2 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 2.6 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 1.6 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.1 | 1.0 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 2.9 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 0.7 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 9.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 3.8 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 1.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 2.7 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 3.3 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 4.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.5 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.3 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 0.4 | GO:0045252 | dihydrolipoyl dehydrogenase complex(GO:0045240) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.5 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 5.2 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 0.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 2.3 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 0.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 2.5 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 3.3 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 1.0 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 12.2 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.5 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 10.9 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.4 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 2.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 3.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 18.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 6.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.8 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 2.9 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 3.5 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.8 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 1.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 7.8 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 2.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.3 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 1.1 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 4.2 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 10.2 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 5.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 3.3 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 17.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 1.9 | 7.5 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 1.8 | 5.3 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 1.6 | 4.8 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 1.5 | 9.1 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 1.5 | 4.5 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 1.1 | 22.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 1.1 | 3.2 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 1.0 | 4.2 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.9 | 4.7 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.9 | 16.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.9 | 3.5 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.9 | 2.6 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.9 | 2.6 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.9 | 3.4 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.7 | 2.2 | GO:0034512 | box C/D snoRNA binding(GO:0034512) |
| 0.7 | 2.9 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.7 | 2.9 | GO:0033265 | choline binding(GO:0033265) |
| 0.7 | 2.1 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.7 | 2.1 | GO:0003860 | 3-hydroxyisobutyryl-CoA hydrolase activity(GO:0003860) |
| 0.7 | 4.0 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.6 | 2.6 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.6 | 9.5 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.6 | 5.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.6 | 2.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.6 | 4.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.5 | 3.2 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.5 | 5.9 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.5 | 2.8 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.5 | 2.3 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.5 | 5.1 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.5 | 2.3 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.4 | 1.3 | GO:0002135 | CTP binding(GO:0002135) |
| 0.4 | 12.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.4 | 5.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.4 | 5.9 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.4 | 1.2 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.4 | 2.8 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.4 | 3.2 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.4 | 3.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.4 | 3.5 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.4 | 7.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.3 | 5.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.3 | 2.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.3 | 2.9 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.3 | 19.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.3 | 0.9 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.3 | 1.5 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.3 | 4.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.3 | 2.6 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.3 | 4.5 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.3 | 9.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.3 | 1.4 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.3 | 2.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.3 | 2.6 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.3 | 0.8 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.3 | 11.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.3 | 4.7 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.3 | 2.5 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.2 | 1.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.2 | 0.7 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.2 | 6.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 6.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 1.3 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.2 | 1.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 2.3 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.2 | 1.6 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.2 | 2.7 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.2 | 0.6 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.2 | 0.8 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 0.6 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.2 | 2.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 4.6 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.2 | 1.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.2 | 9.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.2 | 2.2 | GO:0047617 | CoA hydrolase activity(GO:0016289) acyl-CoA hydrolase activity(GO:0047617) |
| 0.2 | 2.8 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.2 | 1.7 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.1 | 1.6 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 7.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 6.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 2.6 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 1.5 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 2.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 2.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 0.4 | GO:0052854 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 0.1 | 3.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.7 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 1.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.5 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 2.2 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 0.2 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.1 | 0.4 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.4 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 1.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 1.7 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.3 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.1 | 0.3 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 1.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 2.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 2.4 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 2.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.6 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.6 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.1 | 2.0 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 3.8 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 2.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.4 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 1.6 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 3.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 1.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 2.0 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 1.9 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.9 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 5.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 2.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.3 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 0.5 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.9 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 1.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.1 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 2.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.5 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.4 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.4 | GO:0070492 | sialic acid binding(GO:0033691) oligosaccharide binding(GO:0070492) |
| 0.0 | 0.9 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.3 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 8.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 12.9 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.8 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 1.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.3 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.6 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 2.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.5 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 1.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.4 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 1.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 2.1 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 3.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.8 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 1.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.4 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 1.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 2.6 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 1.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.6 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 2.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.0 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 8.4 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.1 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 0.5 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 4.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 2.9 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 2.6 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 9.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 8.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 25.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.2 | 6.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 3.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 2.0 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 10.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 5.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 17.4 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 2.3 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 3.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 5.2 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 6.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 1.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 1.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 2.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 1.8 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 1.8 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 6.8 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 0.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 3.0 | ST ADRENERGIC | Adrenergic Pathway |
| 0.1 | 1.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 1.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 5.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 1.8 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.7 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 2.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 2.0 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 2.6 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 2.3 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.2 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 2.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 4.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.5 | 19.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.5 | 5.8 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.5 | 9.5 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.4 | 8.8 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.3 | 27.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.3 | 10.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 3.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 6.5 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 6.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 15.8 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.2 | 6.9 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.2 | 1.8 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.2 | 5.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 1.5 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.2 | 4.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 4.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.2 | 4.1 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.2 | 4.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 2.0 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 6.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 15.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 5.4 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.1 | 2.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 3.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 3.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 4.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 2.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 3.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.0 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 2.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 2.6 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 1.3 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 1.4 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.1 | 2.3 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.1 | 4.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 2.1 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 2.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 2.2 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.1 | 0.6 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 4.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 12.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 1.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.7 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 1.0 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 6.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 6.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.9 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 3.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 2.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.8 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 12.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 2.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.6 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.1 | 2.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 5.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 1.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 2.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 4.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 2.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 2.4 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.9 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 1.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.9 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.8 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.4 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 1.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.5 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.7 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.5 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 1.6 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.9 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.1 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |