Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for IRF2_STAT2_IRF8_IRF1
Z-value: 3.49
Transcription factors associated with IRF2_STAT2_IRF8_IRF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
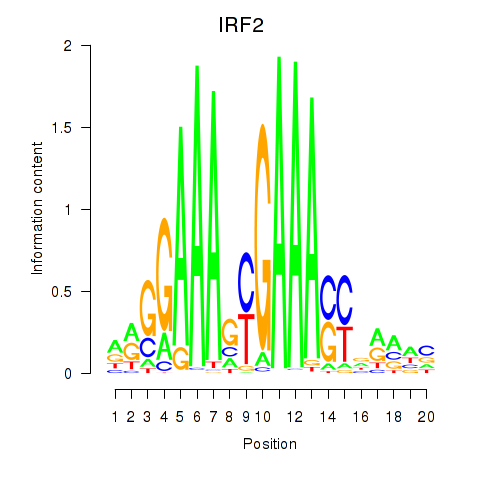
IRF2
|
ENSG00000168310.11 | IRF2 |
|
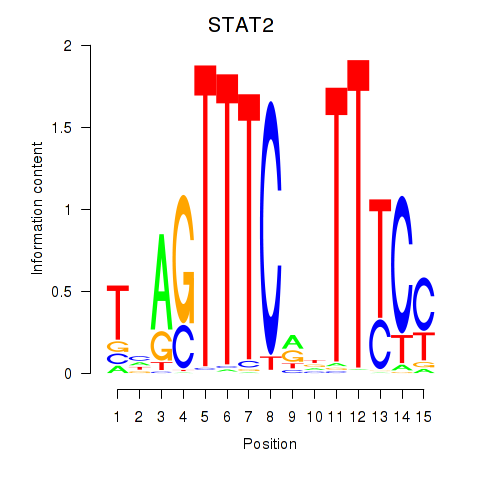
STAT2
|
ENSG00000170581.14 | STAT2 |
|
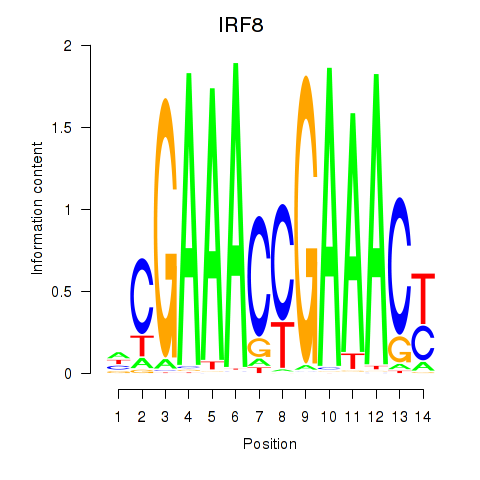
IRF8
|
ENSG00000140968.11 | IRF8 |
|
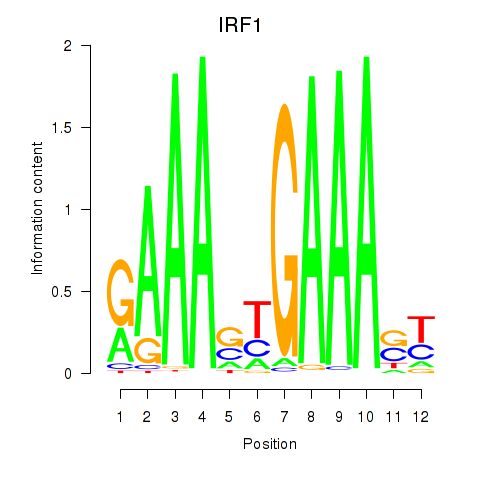
IRF1
|
ENSG00000125347.15 | IRF1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRF1 | hg38_v1_chr5_-_132490750_132490840 | 0.80 | 9.7e-50 | Click! |
| IRF2 | hg38_v1_chr4_-_184474518_184474556 | 0.71 | 1.9e-34 | Click! |
| IRF8 | hg38_v1_chr16_+_85908988_85909033 | 0.61 | 1.7e-23 | Click! |
| STAT2 | hg38_v1_chr12_-_56360084_56360146 | 0.13 | 6.1e-02 | Click! |
Activity profile of IRF2_STAT2_IRF8_IRF1 motif
Sorted Z-values of IRF2_STAT2_IRF8_IRF1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of IRF2_STAT2_IRF8_IRF1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 52.4 | 157.1 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 51.5 | 154.4 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 45.1 | 225.5 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 37.7 | 113.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 33.4 | 267.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 31.4 | 94.2 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 29.5 | 59.0 | GO:0002428 | antigen processing and presentation of peptide antigen via MHC class Ib(GO:0002428) |
| 23.2 | 116.1 | GO:0046103 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 21.8 | 436.4 | GO:0003374 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 19.4 | 154.8 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 19.2 | 96.1 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 18.1 | 180.5 | GO:0018377 | protein myristoylation(GO:0018377) |
| 17.3 | 51.9 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 16.4 | 180.1 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 16.1 | 48.2 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 15.7 | 62.8 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 15.2 | 91.1 | GO:1990668 | vesicle fusion with endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membrane(GO:1990668) |
| 13.8 | 41.5 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 12.9 | 77.3 | GO:0035549 | positive regulation of interferon-beta secretion(GO:0035549) |
| 12.0 | 36.1 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 11.0 | 66.2 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 10.7 | 32.2 | GO:0035915 | pore formation in membrane of other organism(GO:0035915) |
| 10.1 | 40.6 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 9.0 | 27.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 8.9 | 26.6 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 8.7 | 886.1 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 8.6 | 102.9 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 8.5 | 51.2 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 8.3 | 74.4 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 8.2 | 24.5 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 8.2 | 228.3 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 7.8 | 23.4 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 7.8 | 163.9 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 7.5 | 67.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 7.3 | 29.2 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 7.0 | 63.3 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 7.0 | 27.9 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 6.7 | 26.9 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 6.6 | 33.0 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 6.6 | 26.4 | GO:0039516 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 6.5 | 45.5 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 6.3 | 56.8 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 6.3 | 25.0 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 5.6 | 39.5 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 5.0 | 15.0 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 4.7 | 28.1 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 4.6 | 18.5 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 4.6 | 64.1 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 4.5 | 22.4 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 4.3 | 12.8 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 4.2 | 16.7 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 4.2 | 199.2 | GO:0032731 | positive regulation of interleukin-1 beta production(GO:0032731) |
| 4.1 | 33.2 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 4.1 | 20.4 | GO:1902044 | Fas signaling pathway(GO:0036337) regulation of Fas signaling pathway(GO:1902044) |
| 4.0 | 11.9 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 3.8 | 26.7 | GO:0080009 | mRNA methylation(GO:0080009) |
| 3.8 | 34.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 3.8 | 15.2 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 3.7 | 14.7 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 3.6 | 14.5 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 3.6 | 10.7 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 3.4 | 13.6 | GO:0006272 | leading strand elongation(GO:0006272) |
| 3.2 | 9.6 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 3.1 | 12.6 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 3.1 | 18.8 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 3.1 | 55.8 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 3.1 | 36.7 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 3.0 | 21.1 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 3.0 | 14.9 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 2.8 | 11.3 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 2.8 | 11.3 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 2.8 | 8.3 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 2.8 | 13.8 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 2.7 | 16.3 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 2.7 | 16.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 2.5 | 32.5 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 2.5 | 41.8 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 2.4 | 17.0 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 2.4 | 31.5 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 2.4 | 16.7 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 2.3 | 7.0 | GO:2000753 | regulation of phospholipid scramblase activity(GO:1900161) positive regulation of phospholipid scramblase activity(GO:1900163) regulation of glucosylceramide catabolic process(GO:2000752) positive regulation of glucosylceramide catabolic process(GO:2000753) regulation of sphingomyelin catabolic process(GO:2000754) positive regulation of sphingomyelin catabolic process(GO:2000755) |
| 2.3 | 9.3 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 2.3 | 9.2 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 2.3 | 9.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 2.2 | 13.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 2.2 | 99.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 2.1 | 12.9 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 2.1 | 19.2 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 2.1 | 55.8 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 2.0 | 12.2 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 2.0 | 16.0 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 2.0 | 2.0 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 2.0 | 29.4 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 2.0 | 25.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 1.9 | 11.6 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 1.9 | 7.7 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 1.9 | 7.7 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 1.9 | 105.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 1.9 | 7.6 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 1.9 | 7.4 | GO:0002384 | hepatic immune response(GO:0002384) |
| 1.8 | 7.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 1.8 | 1.8 | GO:2000152 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) regulation of ubiquitin-specific protease activity(GO:2000152) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 1.7 | 13.8 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 1.7 | 3.4 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 1.7 | 10.1 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 1.7 | 5.1 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 1.6 | 4.8 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 1.6 | 38.3 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 1.5 | 6.2 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 1.5 | 9.0 | GO:0031437 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 1.5 | 26.5 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 1.4 | 10.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 1.4 | 170.7 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 1.4 | 5.6 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 1.3 | 65.3 | GO:0034383 | low-density lipoprotein particle clearance(GO:0034383) |
| 1.3 | 23.8 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 1.3 | 19.7 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 1.3 | 16.9 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 1.3 | 9.1 | GO:0071351 | interleukin-18-mediated signaling pathway(GO:0035655) cellular response to interleukin-18(GO:0071351) |
| 1.3 | 9.0 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 1.3 | 24.0 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 1.2 | 6.0 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 1.2 | 21.0 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 1.2 | 106.4 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 1.2 | 8.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 1.1 | 8.9 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 1.1 | 5.5 | GO:1904764 | vacuolar transmembrane transport(GO:0034486) chaperone-mediated protein transport involved in chaperone-mediated autophagy(GO:0061741) negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 1.1 | 2.2 | GO:0034756 | regulation of iron ion transport(GO:0034756) regulation of iron ion transmembrane transport(GO:0034759) |
| 1.1 | 16.6 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 1.1 | 8.8 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 1.1 | 3.3 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) negative regulation of mitotic cell cycle DNA replication(GO:1903464) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 1.1 | 4.4 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 1.1 | 3.3 | GO:0061386 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 1.1 | 43.9 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 1.1 | 2.1 | GO:0071283 | cellular response to iron(III) ion(GO:0071283) |
| 1.1 | 53.8 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 1.1 | 4.2 | GO:0006581 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 1.0 | 5.2 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 1.0 | 38.7 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 1.0 | 7.3 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 1.0 | 2.1 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 1.0 | 4.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 1.0 | 3.0 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 1.0 | 4.0 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 1.0 | 6.0 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 1.0 | 144.3 | GO:0015992 | proton transport(GO:0015992) |
| 1.0 | 11.7 | GO:0001675 | acrosome assembly(GO:0001675) |
| 1.0 | 4.8 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 1.0 | 4.8 | GO:0035566 | regulation of metanephros size(GO:0035566) |
| 0.9 | 4.7 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.9 | 15.0 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.9 | 12.1 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.9 | 6.4 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.9 | 2.7 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.9 | 4.4 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.9 | 2.7 | GO:0009720 | detection of hormone stimulus(GO:0009720) |
| 0.9 | 24.6 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.9 | 6.1 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.9 | 5.2 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.9 | 18.0 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.9 | 11.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.9 | 12.8 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.8 | 11.7 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.8 | 13.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.8 | 35.8 | GO:0002456 | T cell mediated immunity(GO:0002456) |
| 0.8 | 6.6 | GO:0015886 | heme transport(GO:0015886) |
| 0.8 | 9.7 | GO:0034138 | toll-like receptor 3 signaling pathway(GO:0034138) |
| 0.8 | 6.3 | GO:0002353 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.8 | 19.7 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.8 | 7.0 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.8 | 7.0 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.8 | 4.5 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.7 | 10.9 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.7 | 2.1 | GO:0090309 | regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.7 | 0.7 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.7 | 2.1 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.7 | 20.4 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.7 | 4.8 | GO:0046449 | creatinine metabolic process(GO:0046449) |
| 0.7 | 2.7 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.7 | 2.0 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.7 | 19.0 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.7 | 16.3 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.6 | 2.6 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.6 | 7.6 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.6 | 2.4 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.6 | 1.2 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.6 | 1.8 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.6 | 1.8 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.6 | 31.8 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.6 | 1.1 | GO:2000525 | negative regulation of regulatory T cell differentiation(GO:0045590) regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.6 | 3.4 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.6 | 3.9 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.5 | 5.5 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.5 | 12.0 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.5 | 11.4 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.5 | 1.6 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.5 | 4.7 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.5 | 6.2 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.5 | 6.0 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.5 | 1.0 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.5 | 10.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.5 | 1.0 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) regulation of forebrain neuron differentiation(GO:2000977) |
| 0.5 | 0.5 | GO:1990791 | dorsal root ganglion development(GO:1990791) |
| 0.5 | 1.9 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.5 | 9.2 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.5 | 4.5 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.4 | 3.1 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.4 | 3.1 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.4 | 5.6 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.4 | 2.1 | GO:0090385 | phagosome acidification(GO:0090383) phagosome-lysosome fusion(GO:0090385) |
| 0.4 | 0.8 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.4 | 26.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.4 | 2.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.4 | 2.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.4 | 5.9 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.4 | 5.7 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.4 | 3.8 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.4 | 14.3 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.4 | 2.6 | GO:0032480 | negative regulation of type I interferon production(GO:0032480) |
| 0.4 | 0.7 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.4 | 2.9 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.4 | 1.4 | GO:0032916 | positive regulation of transforming growth factor beta3 production(GO:0032916) chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.4 | 3.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.3 | 2.1 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.3 | 2.4 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.3 | 0.7 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.3 | 4.4 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.3 | 14.9 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.3 | 1.6 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.3 | 3.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.3 | 2.1 | GO:0015866 | ADP transport(GO:0015866) |
| 0.3 | 10.0 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.3 | 12.5 | GO:0031640 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.3 | 13.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.3 | 20.6 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.3 | 4.6 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.3 | 0.9 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.3 | 5.3 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.3 | 4.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.3 | 1.6 | GO:0051344 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.3 | 2.1 | GO:0045901 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.3 | 38.0 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.3 | 9.9 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.3 | 5.7 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.3 | 1.3 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.2 | 2.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.2 | 6.4 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.2 | 12.4 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.2 | 40.6 | GO:0038096 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.2 | 1.4 | GO:0015755 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.2 | 2.9 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.2 | 9.8 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.2 | 2.6 | GO:0046149 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.2 | 7.5 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.2 | 3.7 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.2 | 1.8 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.2 | 0.9 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.2 | 0.9 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.2 | 2.3 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.2 | 3.6 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 | 3.9 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.2 | 3.0 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.2 | 0.4 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.2 | 11.1 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.2 | 1.0 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.2 | 10.7 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.2 | 18.7 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.2 | 2.0 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.2 | 2.4 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.2 | 14.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.2 | 4.8 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.2 | 28.8 | GO:0071774 | response to fibroblast growth factor(GO:0071774) |
| 0.2 | 11.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.2 | 5.0 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.2 | 3.6 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.2 | 94.4 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.2 | 14.8 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.2 | 4.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 2.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 7.2 | GO:0048477 | oogenesis(GO:0048477) |
| 0.2 | 1.3 | GO:0035989 | tendon development(GO:0035989) |
| 0.2 | 2.7 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.2 | 0.8 | GO:0021764 | amygdala development(GO:0021764) |
| 0.2 | 0.5 | GO:1903412 | response to bile acid(GO:1903412) cellular response to bile acid(GO:1903413) |
| 0.1 | 2.4 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 9.3 | GO:0002200 | somatic diversification of immune receptors(GO:0002200) |
| 0.1 | 2.8 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.1 | 9.2 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 30.2 | GO:0009615 | response to virus(GO:0009615) |
| 0.1 | 19.7 | GO:0006310 | DNA recombination(GO:0006310) |
| 0.1 | 0.8 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.1 | 0.4 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.1 | 6.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 4.1 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 2.1 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 0.6 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 4.9 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 | 1.7 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.7 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 4.3 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.1 | 0.9 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 0.7 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) histone H2A K63-linked ubiquitination(GO:0070535) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 1.5 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.3 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.1 | 4.7 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.1 | 4.2 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 2.5 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.4 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 3.8 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.1 | 1.3 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 2.9 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.1 | 0.8 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 5.3 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.1 | 0.3 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.7 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 1.1 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 | 0.2 | GO:1904075 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.0 | 1.6 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 2.6 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 3.6 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.0 | 1.9 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 1.3 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 2.0 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.3 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.0 | 0.6 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 1.7 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.2 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.0 | 1.2 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.1 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.8 | GO:0007286 | spermatid development(GO:0007286) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 75.9 | 531.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 49.6 | 248.2 | GO:0042825 | TAP complex(GO:0042825) |
| 23.5 | 117.6 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 20.7 | 82.9 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 12.8 | 51.4 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 11.8 | 59.0 | GO:0031905 | early endosome lumen(GO:0031905) |
| 11.2 | 67.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 10.2 | 30.5 | GO:0044393 | microspike(GO:0044393) |
| 9.4 | 47.0 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 9.2 | 202.2 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 8.9 | 80.4 | GO:0032010 | phagolysosome(GO:0032010) |
| 6.9 | 82.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 6.8 | 47.6 | GO:0043196 | varicosity(GO:0043196) |
| 6.3 | 25.0 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 5.8 | 23.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 5.6 | 39.1 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 5.4 | 37.7 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 5.0 | 14.9 | GO:0071756 | IgM immunoglobulin complex(GO:0071753) IgM immunoglobulin complex, circulating(GO:0071754) pentameric IgM immunoglobulin complex(GO:0071756) |
| 4.5 | 40.6 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 4.5 | 40.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 4.5 | 13.4 | GO:0036398 | TCR signalosome(GO:0036398) |
| 4.4 | 57.0 | GO:0005688 | U6 snRNP(GO:0005688) |
| 4.0 | 24.0 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 3.9 | 27.4 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 3.6 | 10.7 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 3.4 | 50.3 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 3.2 | 9.6 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 3.0 | 23.6 | GO:0072487 | MSL complex(GO:0072487) |
| 2.9 | 202.5 | GO:0015030 | Cajal body(GO:0015030) |
| 2.9 | 11.7 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 2.7 | 13.6 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 2.7 | 51.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 2.7 | 29.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 2.6 | 23.6 | GO:0097452 | GAIT complex(GO:0097452) |
| 2.2 | 6.7 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 2.0 | 264.5 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 1.9 | 19.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 1.8 | 32.0 | GO:0005840 | ribosome(GO:0005840) |
| 1.8 | 7.0 | GO:0044307 | dendritic branch(GO:0044307) |
| 1.6 | 12.5 | GO:0044754 | autolysosome(GO:0044754) |
| 1.5 | 13.7 | GO:0042382 | paraspeckles(GO:0042382) |
| 1.5 | 11.9 | GO:0005827 | polar microtubule(GO:0005827) |
| 1.5 | 7.4 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 1.5 | 180.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 1.3 | 5.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 1.3 | 7.7 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 1.3 | 9.0 | GO:0016589 | NURF complex(GO:0016589) |
| 1.3 | 80.4 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 1.3 | 10.0 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 1.2 | 6.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 1.2 | 3.7 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 1.2 | 20.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 1.2 | 17.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 1.2 | 22.4 | GO:0098553 | integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 1.2 | 12.8 | GO:0032039 | integrator complex(GO:0032039) |
| 1.2 | 12.7 | GO:0030870 | Mre11 complex(GO:0030870) |
| 1.2 | 38.0 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 1.2 | 458.9 | GO:0031965 | nuclear membrane(GO:0031965) |
| 1.1 | 53.8 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 1.1 | 5.5 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 1.1 | 4.2 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 1.0 | 5.2 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 1.0 | 3.0 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.8 | 7.6 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.8 | 6.6 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.8 | 10.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.8 | 7.4 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.8 | 4.0 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.8 | 3.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.7 | 25.1 | GO:0016514 | SWI/SNF complex(GO:0016514) npBAF complex(GO:0071564) |
| 0.7 | 9.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.7 | 23.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.7 | 19.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.7 | 4.1 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.7 | 3.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.6 | 10.2 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.6 | 17.4 | GO:0043657 | host(GO:0018995) host cell(GO:0043657) |
| 0.6 | 1.8 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.6 | 6.4 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.6 | 16.7 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.6 | 5.6 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.5 | 6.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.5 | 6.0 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.5 | 24.5 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.5 | 229.8 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.5 | 10.4 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.5 | 6.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.5 | 13.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.4 | 5.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.4 | 28.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.4 | 6.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.4 | 13.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.4 | 20.9 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.4 | 2.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.4 | 2.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.4 | 14.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.4 | 7.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.4 | 10.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.4 | 7.8 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.4 | 16.3 | GO:0032838 | cell projection cytoplasm(GO:0032838) axon cytoplasm(GO:1904115) |
| 0.3 | 17.9 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.3 | 15.9 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.3 | 4.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.3 | 4.9 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.3 | 20.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.3 | 33.2 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.3 | 2.9 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.3 | 12.6 | GO:0005776 | autophagosome(GO:0005776) |
| 0.3 | 44.5 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.3 | 1.4 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.3 | 4.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.3 | 0.8 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.3 | 16.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.3 | 4.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.3 | 47.5 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.2 | 9.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.2 | 20.0 | GO:0005814 | centriole(GO:0005814) |
| 0.2 | 121.8 | GO:0005730 | nucleolus(GO:0005730) |
| 0.2 | 5.9 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.2 | 58.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.2 | 34.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.2 | 75.6 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.2 | 22.0 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.2 | 33.3 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.2 | 2.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.2 | 669.5 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.2 | 7.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 12.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.2 | 12.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 1.9 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.2 | 1.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 7.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 1.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 180.6 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.1 | 10.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 6.7 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.1 | 5.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 2.0 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 0.7 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 23.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 0.6 | GO:0032059 | bleb(GO:0032059) |
| 0.1 | 3.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 30.5 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 7.4 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.1 | 4.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 4.8 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 1.1 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 2.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 2.2 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.2 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.7 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 42.1 | 252.5 | GO:0046979 | TAP2 binding(GO:0046979) |
| 37.7 | 113.2 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 25.5 | 152.8 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 22.3 | 623.8 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 16.4 | 213.5 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 16.2 | 48.6 | GO:0048030 | disaccharide binding(GO:0048030) |
| 15.8 | 47.5 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 13.7 | 68.4 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 11.8 | 82.9 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 11.4 | 102.7 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 11.0 | 55.0 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 10.9 | 151.9 | GO:0031386 | protein tag(GO:0031386) |
| 10.9 | 54.3 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 10.3 | 61.9 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 10.1 | 80.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 9.2 | 275.8 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 9.1 | 145.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 8.1 | 48.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 7.8 | 23.4 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 6.8 | 34.2 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 6.8 | 40.6 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 5.8 | 17.5 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 5.3 | 26.7 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 5.2 | 36.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 5.0 | 15.1 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 4.8 | 24.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 4.8 | 23.8 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 4.5 | 13.6 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 4.5 | 99.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 4.3 | 17.0 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 4.1 | 16.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 4.0 | 24.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 4.0 | 16.0 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 4.0 | 11.9 | GO:0031208 | POZ domain binding(GO:0031208) |
| 4.0 | 19.8 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 3.8 | 103.1 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 3.8 | 15.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 3.7 | 14.9 | GO:0005151 | interleukin-1, Type II receptor binding(GO:0005151) |
| 3.3 | 16.7 | GO:0008948 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) oxaloacetate decarboxylase activity(GO:0008948) |
| 3.3 | 13.4 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 3.2 | 12.9 | GO:0043273 | CTPase activity(GO:0043273) |
| 3.1 | 24.9 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 3.1 | 9.2 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 3.0 | 9.1 | GO:0042008 | interleukin-18 receptor activity(GO:0042008) |
| 3.0 | 36.1 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 2.9 | 108.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 2.8 | 11.3 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 2.8 | 19.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 2.7 | 70.8 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 2.7 | 24.0 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 2.6 | 7.9 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 2.6 | 117.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 2.6 | 79.7 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 2.5 | 7.4 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 2.5 | 41.9 | GO:0019864 | IgG binding(GO:0019864) |
| 2.3 | 27.5 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 2.2 | 11.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 2.2 | 15.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 2.1 | 32.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 2.1 | 8.5 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 2.1 | 35.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 2.1 | 27.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 2.0 | 90.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 2.0 | 6.0 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 2.0 | 31.4 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 1.9 | 7.6 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 1.9 | 3.8 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 1.8 | 5.5 | GO:0047756 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) chondroitin 4-sulfotransferase activity(GO:0047756) |
| 1.8 | 50.9 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 1.7 | 12.2 | GO:0019863 | IgE binding(GO:0019863) |
| 1.7 | 20.4 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 1.7 | 13.6 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 1.7 | 205.0 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 1.7 | 28.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 1.6 | 4.9 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 1.6 | 14.7 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 1.6 | 6.4 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 1.5 | 6.2 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 1.4 | 4.3 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 1.4 | 7.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 1.4 | 5.6 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 1.4 | 6.8 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 1.3 | 9.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 1.3 | 3.9 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 1.3 | 29.7 | GO:0044548 | S100 protein binding(GO:0044548) |
| 1.3 | 232.8 | GO:0001047 | core promoter binding(GO:0001047) |
| 1.3 | 9.0 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 1.3 | 24.0 | GO:0051400 | BH domain binding(GO:0051400) |
| 1.2 | 438.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 1.2 | 36.1 | GO:0001968 | fibronectin binding(GO:0001968) |
| 1.1 | 11.9 | GO:0046790 | virion binding(GO:0046790) |
| 1.1 | 6.5 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 1.1 | 9.6 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 1.1 | 40.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 1.1 | 7.4 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 1.0 | 54.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 1.0 | 7.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 1.0 | 12.0 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 1.0 | 19.8 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 1.0 | 14.8 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 1.0 | 20.6 | GO:0001093 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) TFIIB-class transcription factor binding(GO:0001093) |
| 1.0 | 22.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 1.0 | 19.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 1.0 | 9.7 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 1.0 | 27.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.9 | 7.6 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.9 | 15.0 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.9 | 82.5 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.9 | 32.6 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.9 | 6.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.9 | 14.5 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.9 | 2.7 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.9 | 18.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.9 | 43.9 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.9 | 5.1 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.8 | 6.6 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.8 | 31.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.8 | 13.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.7 | 6.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.7 | 2.2 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.7 | 32.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.7 | 6.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.7 | 8.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.7 | 2.6 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.6 | 107.8 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.6 | 10.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.6 | 3.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.6 | 11.6 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.6 | 2.4 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.6 | 35.7 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.6 | 3.9 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.5 | 14.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.5 | 4.7 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.5 | 9.2 | GO:0004407 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.5 | 21.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.4 | 4.9 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.4 | 2.2 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.4 | 1.7 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.4 | 13.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.4 | 9.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.4 | 26.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.4 | 17.1 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.4 | 1.6 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.4 | 5.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.4 | 6.2 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.4 | 5.7 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.4 | 3.4 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.4 | 28.6 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.4 | 5.7 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.4 | 2.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.3 | 2.0 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.3 | 197.6 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.3 | 1.9 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.3 | 15.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.3 | 31.4 | GO:0005525 | GTP binding(GO:0005525) |
| 0.3 | 8.4 | GO:0048038 | quinone binding(GO:0048038) |
| 0.3 | 9.0 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.3 | 5.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.3 | 0.9 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.3 | 2.1 | GO:0005347 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) ATP transmembrane transporter activity(GO:0005347) purine nucleotide transmembrane transporter activity(GO:0015216) ADP transmembrane transporter activity(GO:0015217) |
| 0.3 | 33.3 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.3 | 4.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.3 | 3.6 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.3 | 1.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.3 | 1.6 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.3 | 4.8 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.3 | 56.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.3 | 4.4 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.3 | 3.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.3 | 5.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.3 | 1.3 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.3 | 3.5 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.3 | 21.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.2 | 4.9 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 1.4 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.2 | 4.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 9.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.2 | 4.1 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.2 | 1.8 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.2 | 2.5 | GO:0016208 | AMP binding(GO:0016208) |
| 0.2 | 0.6 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.2 | 4.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.2 | 2.7 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 48.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.2 | 4.5 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.2 | 4.8 | GO:0001848 | complement binding(GO:0001848) |
| 0.2 | 1.3 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.2 | 50.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.2 | 8.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 4.9 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 2.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.2 | 13.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.2 | 2.4 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.2 | 3.9 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.2 | 3.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.7 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 3.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.7 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 3.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 5.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 5.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 11.0 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 2.6 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 2.5 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 23.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 2.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 1.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 5.4 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.1 | 36.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 4.4 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.1 | 2.9 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 1.8 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 1.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 1.9 | GO:0046961 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 8.6 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.1 | 0.6 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.1 | 1.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.7 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 0.3 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.1 | 2.5 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 1.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 1.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 1.0 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 1.0 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:0015087 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 1.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.7 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 1.0 | GO:0032947 | protein complex scaffold(GO:0032947) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.6 | 209.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 4.2 | 58.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 2.7 | 139.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 2.7 | 67.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 2.4 | 66.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 2.4 | 213.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 2.3 | 51.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 2.1 | 141.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 1.8 | 280.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 1.5 | 15.0 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 1.1 | 43.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 1.1 | 205.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 1.0 | 25.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 1.0 | 70.7 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.9 | 35.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.9 | 25.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.9 | 155.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.8 | 46.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.8 | 37.7 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.7 | 15.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.7 | 13.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.7 | 26.0 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.7 | 41.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.7 | 31.0 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.6 | 2.9 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.6 | 21.6 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.5 | 19.2 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.5 | 9.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.5 | 24.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.5 | 4.2 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.5 | 14.0 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.4 | 6.0 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.4 | 12.1 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.4 | 35.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.3 | 5.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.3 | 10.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.3 | 12.8 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.2 | 8.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.2 | 3.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.2 | 4.8 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.2 | 10.3 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.2 | 9.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 14.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 8.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 2.0 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.1 | 4.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 3.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 4.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 1.6 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 1.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 20.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.0 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 2.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.0 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.2 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.6 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 22.6 | 225.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 19.0 | 95.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 16.1 | 48.2 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 13.2 | 1422.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 10.2 | 757.0 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 8.3 | 266.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 6.8 | 67.9 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 5.6 | 101.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 5.4 | 260.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 2.8 | 50.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 2.6 | 57.3 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 2.3 | 43.9 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 2.1 | 35.8 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 2.0 | 62.7 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 1.6 | 129.8 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 1.5 | 38.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 1.5 | 17.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 1.3 | 9.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 1.1 | 16.9 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 1.0 | 15.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 1.0 | 23.3 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 1.0 | 4.8 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.9 | 41.9 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.9 | 42.0 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.9 | 11.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.8 | 4.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.8 | 7.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.8 | 6.9 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.7 | 24.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.7 | 36.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.7 | 29.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.7 | 3.3 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.6 | 45.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.6 | 14.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.6 | 13.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.6 | 48.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.6 | 34.9 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.6 | 50.4 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.5 | 3.4 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.5 | 5.7 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.5 | 5.5 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.5 | 10.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.4 | 12.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.4 | 55.1 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.4 | 10.4 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.4 | 10.7 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.4 | 25.8 | REACTOME INTERFERON SIGNALING | Genes involved in Interferon Signaling |
| 0.4 | 4.5 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.4 | 5.7 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.3 | 8.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.3 | 10.6 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.3 | 21.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.3 | 8.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.3 | 13.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.3 | 11.9 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.3 | 27.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.3 | 8.4 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.3 | 46.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.3 | 3.0 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 11.4 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.2 | 13.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.2 | 6.9 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.2 | 2.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 4.7 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 3.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 20.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.2 | 7.8 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.2 | 1.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.2 | 3.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.2 | 20.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.2 | 6.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 8.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 2.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 2.7 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 6.4 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.1 | 1.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 2.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 0.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.3 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.1 | 4.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 2.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.9 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 4.3 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.1 | 1.0 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.1 | 2.1 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 5.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.9 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.1 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |