Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for IRX3
Z-value: 1.36
Transcription factors associated with IRX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRX3
|
ENSG00000177508.12 | IRX3 |
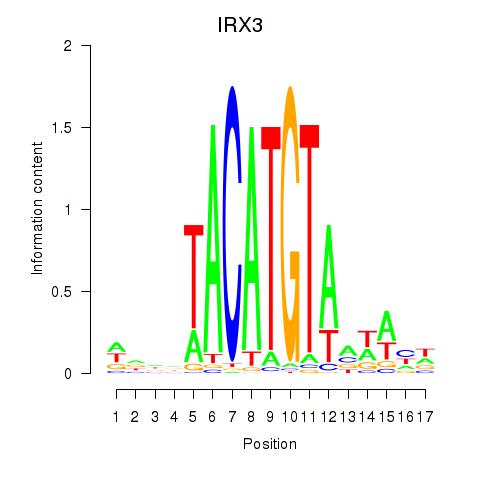
Activity profile of IRX3 motif
Sorted Z-values of IRX3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of IRX3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.6 | 189.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 5.6 | 16.7 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 5.5 | 16.5 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 4.4 | 13.2 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 3.9 | 19.4 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 3.5 | 17.4 | GO:0090131 | mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) mesenchyme migration(GO:0090131) |
| 3.3 | 16.5 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 2.9 | 8.8 | GO:0002884 | negative regulation of hypersensitivity(GO:0002884) |
| 2.9 | 20.4 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 2.5 | 12.3 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 2.4 | 9.4 | GO:0033594 | response to hydroxyisoflavone(GO:0033594) |
| 2.2 | 8.9 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 2.2 | 6.7 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 2.2 | 6.5 | GO:2000866 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 2.1 | 6.4 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 2.1 | 10.5 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 2.1 | 6.3 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 2.0 | 5.9 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 1.9 | 7.7 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 1.9 | 11.4 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 1.9 | 24.5 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 1.8 | 5.4 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 1.7 | 3.4 | GO:1900239 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 1.7 | 13.3 | GO:1903960 | negative regulation of anion transmembrane transport(GO:1903960) |
| 1.7 | 5.0 | GO:1903487 | regulation of lactation(GO:1903487) |
| 1.7 | 26.5 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 1.6 | 16.2 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 1.6 | 17.6 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 1.6 | 12.5 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 1.5 | 21.7 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 1.5 | 4.6 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 1.5 | 4.4 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 1.5 | 8.7 | GO:1904306 | regulation of gastro-intestinal system smooth muscle contraction(GO:1904304) positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 1.4 | 5.5 | GO:1902159 | regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 1.4 | 14.9 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 1.3 | 9.4 | GO:0007000 | nucleolus organization(GO:0007000) |
| 1.3 | 6.4 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 1.3 | 3.8 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 1.2 | 3.7 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 1.2 | 5.8 | GO:0042320 | negative regulation of urine volume(GO:0035811) regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) |
| 1.2 | 3.5 | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 1.1 | 4.5 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 1.1 | 3.3 | GO:0032571 | response to vitamin K(GO:0032571) |
| 1.1 | 6.6 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 1.1 | 2.2 | GO:1902573 | positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 1.1 | 4.3 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 1.1 | 43.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 1.0 | 3.1 | GO:0003104 | positive regulation of glomerular filtration(GO:0003104) |
| 1.0 | 16.0 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 1.0 | 2.9 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 1.0 | 4.8 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.9 | 2.8 | GO:1990451 | cellular stress response to acidic pH(GO:1990451) |
| 0.9 | 2.8 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.9 | 8.2 | GO:0086043 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.9 | 68.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.9 | 3.6 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.9 | 5.4 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.9 | 7.0 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 0.9 | 2.6 | GO:2001188 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001188) |
| 0.9 | 2.6 | GO:0051939 | gamma-aminobutyric acid import(GO:0051939) |
| 0.9 | 9.4 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.9 | 2.6 | GO:0018874 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 0.8 | 3.3 | GO:1903633 | regulation of calcium-dependent ATPase activity(GO:1903610) negative regulation of calcium-dependent ATPase activity(GO:1903611) regulation of dUTP diphosphatase activity(GO:1903627) positive regulation of dUTP diphosphatase activity(GO:1903629) negative regulation of aminoacyl-tRNA ligase activity(GO:1903631) regulation of leucine-tRNA ligase activity(GO:1903633) negative regulation of leucine-tRNA ligase activity(GO:1903634) |
| 0.8 | 9.0 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.8 | 1.6 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.8 | 77.5 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.8 | 3.2 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.8 | 80.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.8 | 4.0 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.8 | 3.9 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.8 | 6.9 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.7 | 15.7 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.7 | 3.0 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.7 | 2.2 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.7 | 7.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.7 | 8.7 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.7 | 5.7 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.7 | 12.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.7 | 2.1 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.7 | 3.4 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.7 | 10.1 | GO:0044241 | lipid digestion(GO:0044241) |
| 0.7 | 3.9 | GO:0003051 | angiotensin catabolic process in blood(GO:0002005) angiotensin-mediated drinking behavior(GO:0003051) multicellular organismal protein catabolic process(GO:0044254) protein digestion(GO:0044256) multicellular organismal macromolecule catabolic process(GO:0044266) |
| 0.6 | 4.4 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.6 | 1.2 | GO:0035691 | macrophage migration inhibitory factor signaling pathway(GO:0035691) |
| 0.6 | 6.7 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.6 | 1.8 | GO:0071934 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.6 | 1.8 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.6 | 16.8 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.6 | 9.4 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.5 | 4.2 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.5 | 1.0 | GO:1990418 | response to insulin-like growth factor stimulus(GO:1990418) |
| 0.5 | 15.1 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.5 | 3.5 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.5 | 2.0 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.5 | 2.0 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.5 | 12.1 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.5 | 8.6 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.5 | 3.8 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.5 | 1.4 | GO:0036215 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.5 | 8.5 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.5 | 4.2 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.5 | 3.7 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.5 | 1.8 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) regulation of rRNA processing(GO:2000232) |
| 0.5 | 3.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.5 | 7.7 | GO:0008212 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.4 | 2.7 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.4 | 1.3 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.4 | 3.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.4 | 3.4 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) |
| 0.4 | 12.6 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) |
| 0.4 | 1.6 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 0.4 | 5.3 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.4 | 3.3 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.4 | 1.6 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.4 | 7.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.4 | 6.7 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.4 | 6.9 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.4 | 9.2 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.4 | 7.5 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.4 | 1.5 | GO:0039507 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.4 | 3.7 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.4 | 4.8 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.4 | 1.1 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.4 | 6.8 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.4 | 4.3 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.4 | 9.6 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.3 | 6.6 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.3 | 3.1 | GO:0060754 | regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.3 | 1.4 | GO:0061358 | negative regulation of Wnt protein secretion(GO:0061358) |
| 0.3 | 3.4 | GO:0001787 | natural killer cell proliferation(GO:0001787) |
| 0.3 | 16.6 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.3 | 10.5 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.3 | 1.3 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.3 | 0.6 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.3 | 2.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.3 | 5.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.3 | 4.4 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.3 | 4.3 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.3 | 2.0 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.3 | 6.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.3 | 2.7 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.3 | 3.2 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.3 | 3.9 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.3 | 0.8 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.3 | 2.0 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.3 | 3.3 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.3 | 0.8 | GO:2000366 | cardiac muscle tissue regeneration(GO:0061026) regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.3 | 1.8 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.2 | 2.0 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.2 | 12.5 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.2 | 10.8 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.2 | 0.5 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.2 | 17.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.2 | 1.1 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.2 | 3.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.2 | 2.8 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.2 | 9.2 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.2 | 0.6 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.2 | 1.0 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.2 | 6.3 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.2 | 4.7 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.2 | 4.3 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.2 | 1.7 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.2 | 0.7 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.2 | 4.0 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.2 | 3.4 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.2 | 3.0 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 3.0 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.2 | 3.7 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 5.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.2 | 3.8 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.2 | 5.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.2 | 6.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.2 | 1.0 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.2 | 1.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.2 | 1.3 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.2 | 18.4 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.2 | 6.6 | GO:0050710 | negative regulation of cytokine secretion(GO:0050710) |
| 0.2 | 4.8 | GO:0044364 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.2 | 11.6 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 3.0 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 1.1 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.1 | 2.9 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.1 | 0.4 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 2.6 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 3.7 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.1 | 10.8 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 1.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 6.1 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.1 | 6.4 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.1 | 1.8 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 2.0 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 3.3 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.1 | 2.8 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 14.1 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 0.9 | GO:0071313 | relaxation of smooth muscle(GO:0044557) cellular response to caffeine(GO:0071313) |
| 0.1 | 1.1 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.1 | 3.9 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 1.9 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 7.0 | GO:1990748 | cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 24.1 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.1 | 1.2 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.1 | 11.1 | GO:1904892 | regulation of JAK-STAT cascade(GO:0046425) regulation of STAT cascade(GO:1904892) |
| 0.1 | 3.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 5.1 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 2.7 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 2.0 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.7 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 1.7 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.1 | 0.8 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 0.9 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.1 | 0.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 3.1 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 0.3 | GO:1903936 | cellular response to sodium arsenite(GO:1903936) |
| 0.1 | 5.9 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 1.3 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 | 1.5 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 3.3 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 17.6 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.1 | 1.0 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.1 | 0.3 | GO:0045628 | T-helper 2 cell differentiation(GO:0045064) regulation of T-helper 2 cell differentiation(GO:0045628) |
| 0.1 | 5.1 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.1 | 0.9 | GO:0034391 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.1 | 2.3 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.1 | 4.5 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 8.5 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.1 | 2.0 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.1 | 0.7 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 2.1 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.1 | 0.3 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.9 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.2 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.1 | 1.0 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 0.8 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 2.3 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 2.1 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.7 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.4 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 7.7 | GO:0007565 | female pregnancy(GO:0007565) |
| 0.0 | 0.5 | GO:0014911 | positive regulation of smooth muscle cell migration(GO:0014911) |
| 0.0 | 1.0 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 1.0 | GO:0034383 | low-density lipoprotein particle clearance(GO:0034383) |
| 0.0 | 3.4 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 1.7 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 1.7 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 1.7 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.0 | 0.1 | GO:0001188 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.0 | 0.2 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 1.3 | GO:0001938 | positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.0 | 1.4 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.4 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.4 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 3.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 2.9 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 2.1 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.1 | GO:0046449 | creatinine metabolic process(GO:0046449) |
| 0.0 | 0.1 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.9 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.8 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.2 | GO:0035855 | platelet formation(GO:0030220) megakaryocyte development(GO:0035855) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.1 | 183.5 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 6.7 | 20.0 | GO:0033150 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
| 6.5 | 19.4 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 5.8 | 17.4 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 5.5 | 16.5 | GO:0036030 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 4.0 | 7.9 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 2.9 | 11.7 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 2.4 | 9.4 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 2.3 | 6.9 | GO:0097229 | sperm end piece(GO:0097229) |
| 2.0 | 21.7 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 1.7 | 6.9 | GO:0000801 | central element(GO:0000801) |
| 1.6 | 4.8 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 1.3 | 5.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 1.3 | 16.7 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 1.3 | 8.9 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 1.1 | 3.2 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 1.0 | 47.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 1.0 | 2.9 | GO:0000805 | X chromosome(GO:0000805) |
| 1.0 | 50.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.9 | 10.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.8 | 6.8 | GO:0034464 | BBSome(GO:0034464) |
| 0.8 | 8.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.8 | 3.9 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.7 | 12.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.7 | 2.8 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.7 | 17.8 | GO:0036019 | endolysosome(GO:0036019) |
| 0.7 | 8.2 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.7 | 6.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.7 | 4.6 | GO:0060091 | kinocilium(GO:0060091) |
| 0.6 | 12.6 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.6 | 11.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.6 | 2.4 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.6 | 7.0 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.6 | 7.5 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.5 | 10.8 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.5 | 4.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.5 | 20.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.5 | 9.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.5 | 1.9 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.5 | 5.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.4 | 4.5 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.4 | 8.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.4 | 8.4 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.4 | 8.8 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.4 | 5.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.4 | 2.6 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.4 | 4.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 4.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.3 | 2.8 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.3 | 2.7 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.3 | 4.3 | GO:0043219 | lateral loop(GO:0043219) |
| 0.3 | 8.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.3 | 1.4 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.3 | 0.8 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.2 | 0.7 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.2 | 4.7 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.2 | 23.3 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.2 | 2.8 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 1.6 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 3.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.2 | 5.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.2 | 0.7 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.2 | 2.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.2 | 55.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.2 | 2.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 11.2 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 2.8 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 1.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 1.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 198.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 13.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 3.0 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 2.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 3.0 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 20.5 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 5.0 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 6.3 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 1.8 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 1.5 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.1 | 0.8 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 2.5 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 16.1 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 2.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 1.7 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 6.3 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 10.5 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 3.3 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 15.2 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 1.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.6 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 2.0 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 3.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 2.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.7 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.9 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 1.3 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 90.3 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 1.7 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 19.4 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 3.8 | 19.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 3.5 | 13.8 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 3.0 | 12.1 | GO:0004773 | steryl-sulfatase activity(GO:0004773) |
| 2.9 | 11.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 2.6 | 7.9 | GO:0042806 | amidase activity(GO:0004040) fucose binding(GO:0042806) |
| 2.5 | 7.5 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 2.4 | 7.2 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 2.4 | 205.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 1.8 | 1.8 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 1.8 | 10.5 | GO:0003796 | lysozyme activity(GO:0003796) |
| 1.7 | 15.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 1.7 | 14.9 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 1.6 | 16.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 1.6 | 11.2 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 1.4 | 4.3 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 1.4 | 7.0 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 1.4 | 16.7 | GO:0008430 | selenium binding(GO:0008430) |
| 1.3 | 13.3 | GO:0005549 | odorant binding(GO:0005549) |
| 1.3 | 2.6 | GO:0048030 | disaccharide binding(GO:0048030) |
| 1.3 | 16.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 1.3 | 3.8 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 1.2 | 3.7 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 1.2 | 3.6 | GO:0004507 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 1.1 | 5.7 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 1.1 | 3.4 | GO:0033765 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 1.1 | 4.5 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 1.1 | 5.5 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 1.1 | 10.8 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 1.1 | 4.3 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 1.1 | 4.2 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 1.0 | 4.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 1.0 | 3.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 1.0 | 3.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 1.0 | 7.0 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 1.0 | 3.0 | GO:0015265 | urea channel activity(GO:0015265) |
| 1.0 | 2.9 | GO:0017129 | triglyceride binding(GO:0017129) |
| 1.0 | 4.8 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.9 | 2.8 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.8 | 50.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.8 | 3.3 | GO:0004341 | gluconolactonase activity(GO:0004341) |
| 0.8 | 3.2 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.8 | 3.9 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.8 | 5.4 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.7 | 66.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.7 | 10.6 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.7 | 12.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.7 | 2.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.7 | 2.1 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.7 | 2.0 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.7 | 4.0 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.7 | 5.3 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.7 | 17.7 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.7 | 16.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.6 | 108.0 | GO:0003823 | antigen binding(GO:0003823) |
| 0.6 | 3.6 | GO:0070728 | leucine binding(GO:0070728) |
| 0.6 | 1.8 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.6 | 1.8 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.6 | 4.0 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.6 | 12.9 | GO:0031005 | filamin binding(GO:0031005) |
| 0.5 | 1.6 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 0.5 | 1.6 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.5 | 3.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.5 | 3.1 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.5 | 3.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.5 | 10.7 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.5 | 9.4 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.5 | 7.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.5 | 5.6 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.5 | 1.4 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.5 | 3.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.5 | 4.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.4 | 2.7 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.4 | 26.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.4 | 3.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.4 | 2.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.4 | 2.0 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.4 | 12.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.4 | 3.9 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.4 | 3.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.4 | 3.8 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.4 | 6.4 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.4 | 38.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.4 | 7.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.4 | 2.6 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.4 | 17.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.4 | 6.7 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.4 | 2.2 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.4 | 4.8 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.4 | 11.3 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.3 | 2.1 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.3 | 12.5 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.3 | 1.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.3 | 1.6 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.3 | 2.6 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.3 | 2.9 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.3 | 6.2 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.3 | 2.6 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.3 | 12.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.3 | 1.6 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.3 | 2.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.3 | 10.1 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.3 | 2.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.3 | 9.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.3 | 2.0 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.3 | 3.4 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.3 | 1.3 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.3 | 9.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.3 | 8.2 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.2 | 1.5 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.2 | 5.9 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.2 | 6.7 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.2 | 14.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 7.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.2 | 7.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.2 | 1.5 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.2 | 4.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 4.1 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.2 | 1.6 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.2 | 0.9 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.2 | 5.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 2.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.2 | 1.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.2 | 2.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.2 | 0.6 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.2 | 5.5 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.2 | 1.7 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.2 | 43.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 5.4 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) |
| 0.2 | 3.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 1.1 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.2 | 1.4 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 2.6 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.2 | 7.2 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.2 | 1.3 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.2 | 8.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.2 | 10.1 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.2 | 4.6 | GO:0071949 | FAD binding(GO:0071949) |
| 0.2 | 0.9 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 13.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.6 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.4 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 12.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.7 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 22.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 1.5 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 1.9 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.8 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 3.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.3 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.1 | 3.4 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 3.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 3.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 18.0 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 3.6 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 3.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 2.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.4 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 1.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 6.6 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 2.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 1.0 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 1.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 0.6 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 0.6 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 1.3 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.1 | 4.7 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 2.2 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 1.0 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 2.8 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.1 | 0.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 2.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 3.9 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.1 | 1.1 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 5.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 11.7 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 2.6 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 15.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 4.6 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 10.8 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.5 | GO:0022840 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.0 | 2.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.2 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 1.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 2.0 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 1.7 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 24.4 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 3.5 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.2 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 3.1 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.2 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 3.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 2.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 16.0 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 1.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.2 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.6 | GO:0004497 | monooxygenase activity(GO:0004497) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 189.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 1.1 | 19.3 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.8 | 17.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.5 | 23.0 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.4 | 73.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.4 | 9.8 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.4 | 23.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.4 | 4.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.4 | 9.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.3 | 16.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 88.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.3 | 15.1 | PID MYC PATHWAY | C-MYC pathway |
| 0.3 | 1.4 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 13.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.2 | 4.7 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 5.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.2 | 47.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 6.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 16.0 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.2 | 2.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.2 | 21.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.2 | 4.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.2 | 9.0 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 15.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 6.9 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 1.9 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 12.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 2.8 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 2.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 1.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 5.8 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 2.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 3.1 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 3.7 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 3.9 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 1.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 1.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 2.8 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 3.7 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 4.4 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 1.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 2.0 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 5.3 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 0.8 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 2.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.8 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 1.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 171.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 2.7 | 16.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 1.2 | 10.9 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 1.1 | 12.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 1.1 | 21.7 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.9 | 9.6 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.8 | 12.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.8 | 7.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.7 | 1.5 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.7 | 10.8 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.7 | 27.7 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.7 | 16.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.7 | 60.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.6 | 6.7 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.5 | 17.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.4 | 10.2 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.4 | 5.3 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.4 | 8.8 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.4 | 12.8 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.4 | 6.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.4 | 16.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.3 | 3.0 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.3 | 9.0 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.3 | 6.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.3 | 3.9 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.3 | 8.0 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.3 | 17.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.3 | 6.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.3 | 2.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.3 | 16.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.3 | 29.3 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.2 | 17.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.2 | 4.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 1.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 6.6 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.2 | 7.7 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.2 | 1.9 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.2 | 1.3 | REACTOME SIGNALING BY ILS | Genes involved in Signaling by Interleukins |
| 0.2 | 2.9 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 2.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.2 | 4.6 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.2 | 2.0 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.2 | 4.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 6.0 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 3.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 6.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 2.7 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 5.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 27.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 4.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 5.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.4 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.1 | 2.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 8.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 6.1 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 1.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 4.9 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 2.1 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 6.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 9.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 2.0 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 10.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 5.7 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 1.5 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 4.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 0.7 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 2.8 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 6.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 0.4 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 1.0 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 4.4 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.1 | 0.9 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 1.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 2.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.7 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.0 | 0.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.8 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 1.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |