Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for IRX6_IRX4
Z-value: 0.48
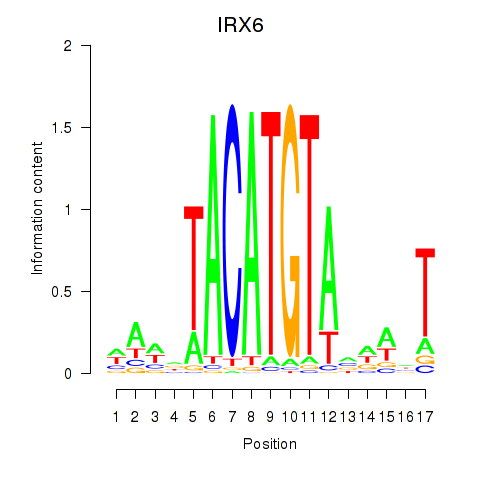
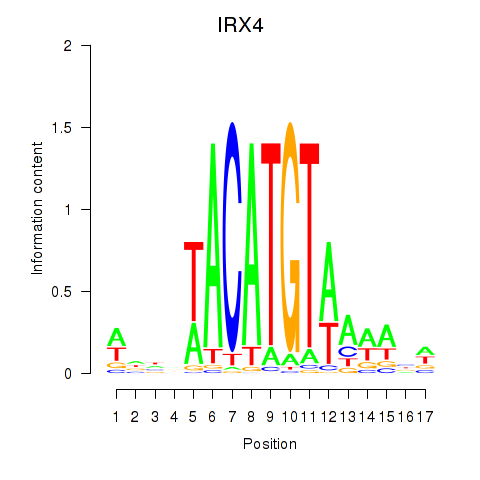
Transcription factors associated with IRX6_IRX4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRX6
|
ENSG00000159387.8 | IRX6 |
|
IRX4
|
ENSG00000113430.10 | IRX4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRX4 | hg38_v1_chr5_-_1886938_1886984, hg38_v1_chr5_-_1882902_1882932 | -0.45 | 4.7e-12 | Click! |
Activity profile of IRX6_IRX4 motif
Sorted Z-values of IRX6_IRX4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of IRX6_IRX4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 1.2 | 15.7 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 1.0 | 6.1 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 1.0 | 19.5 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.9 | 4.5 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.7 | 6.6 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.6 | 3.4 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.5 | 11.0 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.4 | 8.4 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.2 | 3.5 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.2 | 1.0 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.1 | 3.7 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.1 | 1.0 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 2.1 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.1 | 8.1 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 1.8 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 3.0 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 1.5 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.4 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.6 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 2.0 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 7.0 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.6 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 1.4 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.9 | GO:0001578 | microtubule bundle formation(GO:0001578) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 19.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.5 | 6.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.4 | 3.0 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.2 | 8.4 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.2 | 3.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 11.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 8.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.0 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 6.6 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 0.7 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 3.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 2.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 8.1 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.6 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 15.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 3.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 6.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 8.5 | GO:0030529 | intracellular ribonucleoprotein complex(GO:0030529) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.1 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 2.6 | 15.7 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 1.5 | 6.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.7 | 8.4 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.5 | 11.0 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.4 | 3.5 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.2 | 4.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 1.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.7 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.1 | 1.8 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.1 | 2.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 7.0 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 18.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 1.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 3.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 1.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.5 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 4.0 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.6 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 2.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 8.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 11.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 3.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 2.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.0 | PID AURORA A PATHWAY | Aurora A signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 15.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.2 | 3.1 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 8.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 11.9 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.0 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 1.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 1.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |