Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
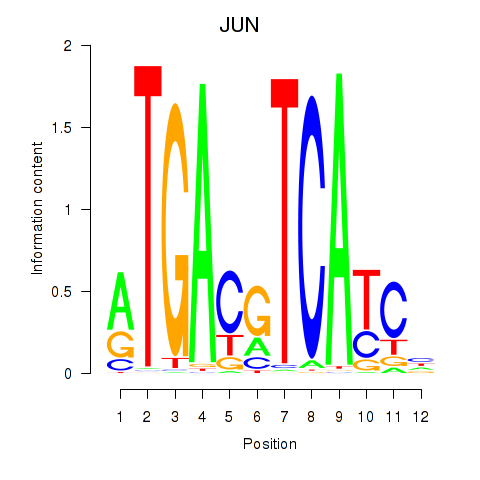
Results for JUN
Z-value: 1.16
Transcription factors associated with JUN
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
JUN
|
ENSG00000177606.8 | JUN |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| JUN | hg38_v1_chr1_-_58784035_58784054 | 0.05 | 4.7e-01 | Click! |
Activity profile of JUN motif
Sorted Z-values of JUN motif
Network of associatons between targets according to the STRING database.
First level regulatory network of JUN
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.0 | 21.1 | GO:1904692 | positive regulation of type B pancreatic cell proliferation(GO:1904692) |
| 6.2 | 18.7 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 5.9 | 17.8 | GO:0061163 | endoplasmic reticulum polarization(GO:0061163) actin filament bundle retrograde transport(GO:0061573) actin filament bundle distribution(GO:0070650) |
| 4.7 | 23.3 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 4.0 | 15.9 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 3.9 | 11.7 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 3.8 | 22.9 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 3.8 | 22.7 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 3.1 | 21.4 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 2.6 | 15.8 | GO:0038016 | insulin receptor internalization(GO:0038016) |
| 2.6 | 23.4 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 2.6 | 10.3 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 2.3 | 6.9 | GO:0046066 | dGDP metabolic process(GO:0046066) |
| 2.2 | 6.7 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 2.1 | 25.2 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 2.1 | 10.3 | GO:0030070 | insulin processing(GO:0030070) |
| 2.0 | 10.2 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 2.0 | 6.0 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 2.0 | 5.9 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 2.0 | 15.8 | GO:0061709 | reticulophagy(GO:0061709) |
| 1.9 | 5.8 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 1.8 | 81.7 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 1.8 | 7.1 | GO:0070426 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 1.8 | 10.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 1.8 | 10.5 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 1.7 | 25.5 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 1.7 | 8.4 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 1.6 | 8.0 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 1.5 | 6.2 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 1.5 | 4.6 | GO:0021897 | forebrain astrocyte differentiation(GO:0021896) forebrain astrocyte development(GO:0021897) |
| 1.5 | 25.7 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 1.5 | 2.9 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 1.4 | 44.4 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 1.4 | 24.4 | GO:0061339 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 1.4 | 4.2 | GO:2000832 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 1.3 | 9.2 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 1.3 | 13.0 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 1.2 | 7.4 | GO:0018343 | protein farnesylation(GO:0018343) |
| 1.2 | 4.8 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 1.2 | 2.4 | GO:0086053 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 1.2 | 3.5 | GO:1903892 | negative regulation of ATF6-mediated unfolded protein response(GO:1903892) |
| 1.2 | 3.5 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 1.2 | 3.5 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 1.2 | 11.6 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.1 | 8.7 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 1.1 | 5.4 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 1.0 | 5.1 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 1.0 | 9.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 1.0 | 17.0 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 1.0 | 8.0 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 1.0 | 2.0 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 1.0 | 4.9 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 1.0 | 13.7 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 1.0 | 11.7 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 1.0 | 8.7 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 1.0 | 16.4 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 1.0 | 3.8 | GO:2001034 | histone H3-K36 dimethylation(GO:0097676) positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.9 | 3.8 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.9 | 3.8 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.9 | 9.1 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.9 | 11.8 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.9 | 10.9 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.9 | 2.7 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.9 | 11.5 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.9 | 6.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.9 | 4.4 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.9 | 17.6 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.9 | 2.6 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.8 | 5.9 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.8 | 5.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.8 | 10.9 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.8 | 24.0 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.8 | 2.5 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.8 | 2.3 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.7 | 1.5 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.7 | 2.9 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.7 | 6.4 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.7 | 5.6 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.7 | 5.6 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.7 | 4.8 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.7 | 10.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.7 | 9.8 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.7 | 14.3 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.6 | 8.9 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.6 | 7.5 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.6 | 1.3 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.6 | 1.9 | GO:0031958 | corticosteroid receptor signaling pathway(GO:0031958) |
| 0.6 | 2.5 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.6 | 12.7 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.6 | 10.2 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.6 | 23.2 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.6 | 5.9 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.6 | 5.3 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.6 | 6.7 | GO:0015871 | choline transport(GO:0015871) |
| 0.5 | 2.2 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.5 | 3.2 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 0.5 | 20.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.5 | 5.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.5 | 9.1 | GO:0071027 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.5 | 2.0 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.5 | 4.5 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.5 | 3.5 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.5 | 1.9 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.5 | 3.9 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.5 | 4.3 | GO:0030822 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) |
| 0.5 | 13.2 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.5 | 1.4 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.5 | 1.4 | GO:1901053 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.5 | 7.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.4 | 5.8 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.4 | 2.6 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.4 | 7.8 | GO:0021895 | cerebral cortex neuron differentiation(GO:0021895) |
| 0.4 | 11.5 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.4 | 4.9 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.4 | 3.2 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.4 | 1.6 | GO:1990637 | hepatic immune response(GO:0002384) response to prolactin(GO:1990637) |
| 0.4 | 21.8 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.4 | 1.6 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.4 | 2.3 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.4 | 0.4 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.4 | 1.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.3 | 2.8 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.3 | 6.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.3 | 1.3 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.3 | 10.4 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.3 | 17.7 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.3 | 10.3 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.3 | 3.9 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.3 | 5.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.3 | 2.9 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.3 | 16.7 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.3 | 1.3 | GO:0046601 | positive regulation of centrosome duplication(GO:0010825) positive regulation of centriole replication(GO:0046601) |
| 0.3 | 16.0 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.3 | 1.2 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.3 | 14.6 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.3 | 0.9 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.3 | 2.6 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.3 | 4.0 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.3 | 0.8 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.3 | 9.5 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.3 | 3.4 | GO:0000050 | urea cycle(GO:0000050) |
| 0.3 | 6.8 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.3 | 3.0 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.2 | 3.0 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.2 | 1.7 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.2 | 3.9 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.2 | 7.6 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.2 | 7.6 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.2 | 15.5 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.2 | 0.9 | GO:0036116 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.2 | 1.6 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.2 | 5.3 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.2 | 6.3 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.2 | 1.1 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.2 | 1.7 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.2 | 1.5 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.2 | 22.4 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.2 | 0.6 | GO:0061386 | soft palate development(GO:0060023) closure of optic fissure(GO:0061386) |
| 0.2 | 20.4 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.2 | 3.9 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.2 | 0.6 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.2 | 1.6 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.2 | 1.6 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.2 | 1.2 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.2 | 29.7 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.2 | 3.3 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.2 | 7.0 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.2 | 1.4 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.2 | 3.3 | GO:1904776 | protein localization to cell cortex(GO:0072697) regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.2 | 7.2 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.2 | 1.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.2 | 1.4 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.2 | 0.9 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 12.6 | GO:0048477 | oogenesis(GO:0048477) |
| 0.2 | 4.8 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.1 | 6.7 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.1 | 1.9 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.1 | 7.2 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 1.0 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.1 | 1.9 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.1 | 2.6 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 5.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 2.2 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.1 | 2.6 | GO:0003094 | glomerular filtration(GO:0003094) renal filtration(GO:0097205) |
| 0.1 | 5.2 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 1.9 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 5.1 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 | 5.2 | GO:0045776 | negative regulation of blood pressure(GO:0045776) |
| 0.1 | 3.9 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 1.5 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 4.4 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.1 | 0.3 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 1.0 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.1 | 0.9 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 7.3 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 5.3 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.1 | 3.7 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 1.3 | GO:0001660 | fever generation(GO:0001660) heat generation(GO:0031649) |
| 0.1 | 9.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 7.5 | GO:0007272 | ensheathment of neurons(GO:0007272) axon ensheathment(GO:0008366) |
| 0.1 | 1.0 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.7 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 2.6 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 4.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 1.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.2 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.1 | 4.4 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.1 | 7.8 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 0.6 | GO:0030813 | positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.0 | 3.8 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 1.8 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 2.9 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.8 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 1.8 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.3 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.2 | GO:0097324 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 1.8 | GO:0006024 | aminoglycan biosynthetic process(GO:0006023) glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 0.4 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.0 | GO:0002876 | positive regulation of chronic inflammatory response to antigenic stimulus(GO:0002876) |
| 0.0 | 0.9 | GO:0035308 | negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.3 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.6 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 1.3 | GO:0007098 | centrosome cycle(GO:0007098) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 23.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 2.9 | 8.7 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 2.4 | 11.9 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 2.3 | 7.0 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 2.2 | 10.9 | GO:0031673 | H zone(GO:0031673) |
| 2.1 | 20.9 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 2.1 | 10.3 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 2.0 | 4.0 | GO:0033011 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
| 1.8 | 7.4 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 1.7 | 23.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 1.3 | 23.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 1.3 | 6.5 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 1.3 | 11.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 1.2 | 20.0 | GO:0008091 | spectrin(GO:0008091) |
| 1.2 | 8.2 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 1.1 | 8.0 | GO:0032021 | NELF complex(GO:0032021) |
| 1.0 | 16.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 1.0 | 22.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 1.0 | 18.3 | GO:0005922 | connexon complex(GO:0005922) |
| 1.0 | 9.0 | GO:0070652 | HAUS complex(GO:0070652) |
| 1.0 | 11.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.8 | 13.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.7 | 2.1 | GO:0044393 | microspike(GO:0044393) |
| 0.7 | 15.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.5 | 18.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.5 | 5.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.5 | 6.5 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.5 | 26.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.4 | 86.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.4 | 1.3 | GO:0098536 | deuterosome(GO:0098536) |
| 0.4 | 5.4 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.4 | 2.7 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.4 | 8.0 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.4 | 4.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.4 | 4.4 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.4 | 33.5 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.3 | 5.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.3 | 1.6 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.3 | 57.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.3 | 4.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.3 | 97.2 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.3 | 1.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.3 | 2.0 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.3 | 18.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.3 | 1.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.3 | 32.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.2 | 17.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 2.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 3.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 9.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.2 | 7.5 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.2 | 2.3 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.2 | 14.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.2 | 2.9 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 3.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.2 | 7.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 3.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 7.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.2 | 5.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 4.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 46.8 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 0.6 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.1 | 11.1 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 9.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 2.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 7.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 1.4 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.1 | 1.3 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 1.0 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 40.6 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.1 | 0.7 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 5.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 6.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 6.0 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 0.7 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 4.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 4.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.3 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 1.7 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.2 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.1 | 4.2 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 16.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 4.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 5.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 4.2 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 1.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 2.8 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 7.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.1 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.8 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 25.7 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 3.1 | 18.3 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 2.9 | 22.9 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 2.8 | 22.7 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 2.6 | 46.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 2.3 | 9.1 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 2.3 | 9.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 2.2 | 8.7 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 2.0 | 18.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 2.0 | 15.8 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 1.9 | 23.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 1.9 | 5.8 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 1.9 | 5.6 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 1.8 | 7.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 1.8 | 25.8 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 1.8 | 7.4 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 1.8 | 25.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 1.7 | 5.1 | GO:0042806 | amidase activity(GO:0004040) fucose binding(GO:0042806) |
| 1.5 | 6.0 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 1.3 | 3.9 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 1.3 | 17.5 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 1.2 | 6.2 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 1.2 | 4.8 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 1.1 | 20.9 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 1.1 | 12.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 1.1 | 5.3 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 1.0 | 8.0 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 1.0 | 5.9 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 1.0 | 9.8 | GO:0042731 | PH domain binding(GO:0042731) |
| 1.0 | 5.7 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.9 | 3.8 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.9 | 4.5 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.9 | 12.5 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.9 | 8.0 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.9 | 3.5 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.9 | 3.4 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.8 | 3.3 | GO:0035473 | lipase binding(GO:0035473) |
| 0.8 | 3.2 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) L-phenylalanine aminotransferase activity(GO:0070546) |
| 0.8 | 3.0 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.7 | 12.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.7 | 3.0 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.7 | 4.4 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.7 | 19.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.7 | 25.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.7 | 6.9 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.7 | 6.8 | GO:0005549 | odorant binding(GO:0005549) |
| 0.7 | 17.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.7 | 2.0 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.6 | 3.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.6 | 1.9 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.6 | 7.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.6 | 53.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.6 | 10.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.6 | 11.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.6 | 1.7 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.5 | 4.4 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.5 | 2.7 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.5 | 11.8 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.5 | 10.5 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.5 | 1.6 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.5 | 27.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.5 | 6.7 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.5 | 3.8 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.5 | 16.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.5 | 8.0 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.5 | 1.4 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.5 | 10.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.4 | 13.0 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.4 | 2.6 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.4 | 70.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.4 | 6.9 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.4 | 10.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.4 | 1.3 | GO:0005135 | interleukin-3 receptor binding(GO:0005135) |
| 0.4 | 10.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.4 | 4.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.4 | 6.0 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.4 | 7.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.4 | 10.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.4 | 13.7 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.4 | 2.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.4 | 7.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.4 | 1.1 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.4 | 4.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.4 | 4.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.3 | 5.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.3 | 1.4 | GO:0034618 | nitric-oxide synthase activity(GO:0004517) arginine binding(GO:0034618) |
| 0.3 | 1.7 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.3 | 19.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.3 | 7.8 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.3 | 1.9 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.3 | 0.6 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.3 | 0.9 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.3 | 4.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.3 | 16.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.3 | 0.8 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.3 | 32.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.3 | 5.0 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 1.5 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.2 | 10.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 5.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.2 | 1.9 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.2 | 1.4 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.2 | 4.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 11.7 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.2 | 3.5 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.2 | 6.3 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.2 | 3.7 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.2 | 1.6 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 72.8 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.2 | 1.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.2 | 7.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 3.6 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.2 | 5.9 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.2 | 1.6 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.2 | 5.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 1.9 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.2 | 1.0 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 3.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 2.9 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 2.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.7 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 10.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 14.0 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.1 | 1.7 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 10.0 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 2.5 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 1.9 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.9 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 9.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 6.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 1.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 1.9 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.1 | 14.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 17.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 3.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.6 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.2 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.1 | 4.5 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 0.3 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.1 | 4.4 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 0.7 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 7.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.3 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.1 | 4.4 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.6 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.9 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 2.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 1.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 11.9 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 15.7 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 3.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.5 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 1.1 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 1.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.4 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 2.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.7 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 4.5 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 1.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 3.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.2 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.9 | GO:0002039 | p53 binding(GO:0002039) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 44.8 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.7 | 17.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.6 | 27.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.5 | 10.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.5 | 16.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.5 | 7.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.4 | 11.7 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.4 | 6.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.3 | 6.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.3 | 4.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.3 | 8.0 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.3 | 14.1 | PID ATM PATHWAY | ATM pathway |
| 0.3 | 15.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.3 | 4.8 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.2 | 7.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.2 | 11.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 11.6 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.2 | 8.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 11.1 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.2 | 4.4 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.2 | 14.7 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.2 | 1.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 1.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 7.4 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.2 | 10.8 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 2.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 11.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 7.4 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 3.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 4.1 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 4.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 8.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 1.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 2.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 2.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 1.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 5.7 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 1.5 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 3.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 1.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 2.5 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 1.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 2.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.9 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.0 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 5.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 46.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 1.6 | 4.9 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 1.5 | 25.7 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 1.2 | 27.4 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 1.1 | 13.0 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 1.0 | 18.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.8 | 34.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.7 | 17.6 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.6 | 13.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.5 | 6.7 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.5 | 10.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.5 | 16.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.4 | 20.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.4 | 15.2 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.4 | 11.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.3 | 9.1 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.3 | 27.1 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.3 | 4.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.3 | 5.0 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.3 | 4.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.3 | 4.8 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 5.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 55.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.2 | 3.9 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 20.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 4.0 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 13.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 7.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 4.3 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.2 | 10.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 7.1 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 1.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 6.7 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.1 | 1.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 4.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 6.2 | REACTOME HEPARAN SULFATE HEPARIN HS GAG METABOLISM | Genes involved in Heparan sulfate/heparin (HS-GAG) metabolism |
| 0.1 | 12.6 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.1 | 1.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 4.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.5 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 1.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 1.6 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 5.2 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 7.8 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 1.1 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 2.0 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 4.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 2.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.7 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 2.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.8 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |