Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for KLF1
Z-value: 1.45
Transcription factors associated with KLF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
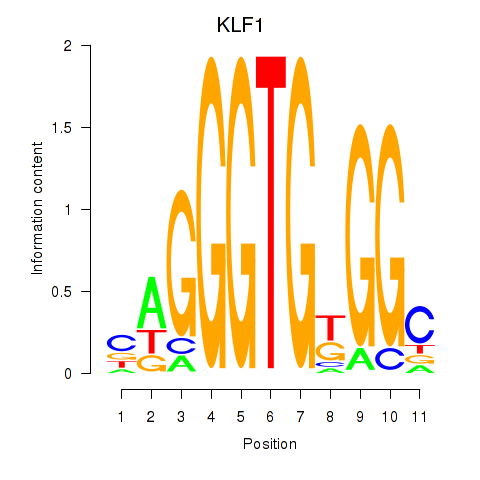
KLF1
|
ENSG00000105610.6 | KLF1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF1 | hg38_v1_chr19_-_12887188_12887207 | -0.30 | 4.6e-06 | Click! |
Activity profile of KLF1 motif
Sorted Z-values of KLF1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.1 | 65.3 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 11.4 | 45.7 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 10.5 | 31.6 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 7.6 | 37.9 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 6.7 | 20.1 | GO:0061445 | endocardial cell fate commitment(GO:0060957) endocardial cushion cell fate commitment(GO:0061445) |
| 6.6 | 26.4 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 4.5 | 18.2 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 4.5 | 44.6 | GO:0002934 | desmosome organization(GO:0002934) |
| 4.2 | 21.1 | GO:1904636 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 3.6 | 10.7 | GO:0021592 | fourth ventricle development(GO:0021592) third ventricle development(GO:0021678) |
| 3.5 | 28.2 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 3.5 | 80.4 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 3.3 | 60.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 3.3 | 19.9 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 3.2 | 12.9 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 3.1 | 9.4 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 3.1 | 3.1 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 2.7 | 10.8 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 2.5 | 10.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 2.5 | 12.3 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 2.4 | 12.1 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 2.2 | 6.6 | GO:0019046 | release from viral latency(GO:0019046) regulation of DNA strand elongation(GO:0060382) |
| 2.2 | 15.3 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 2.2 | 6.5 | GO:1902728 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 2.2 | 30.3 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 2.0 | 8.0 | GO:0036116 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 2.0 | 9.9 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) positive regulation of blood vessel remodeling(GO:2000504) |
| 2.0 | 19.6 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 1.9 | 9.7 | GO:1902896 | terminal web assembly(GO:1902896) |
| 1.9 | 11.3 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 1.9 | 5.6 | GO:0051365 | leading edge cell differentiation(GO:0035026) cellular response to potassium ion starvation(GO:0051365) |
| 1.9 | 18.6 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 1.8 | 22.0 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 1.8 | 29.3 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 1.8 | 12.5 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 1.8 | 19.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 1.7 | 21.9 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 1.6 | 11.5 | GO:0015866 | ADP transport(GO:0015866) |
| 1.6 | 9.8 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 1.5 | 4.5 | GO:0051097 | negative regulation of helicase activity(GO:0051097) oxidative stress-induced premature senescence(GO:0090403) oligodendrocyte apoptotic process(GO:0097252) |
| 1.5 | 10.3 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 1.4 | 11.6 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 1.4 | 17.3 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 1.4 | 18.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 1.4 | 11.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 1.4 | 6.9 | GO:2000542 | negative regulation of gastrulation(GO:2000542) |
| 1.3 | 3.9 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 1.2 | 4.7 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 1.2 | 3.5 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 1.1 | 12.3 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 1.1 | 6.6 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 1.1 | 3.3 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 1.1 | 21.6 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 1.0 | 5.2 | GO:2000814 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 1.0 | 5.1 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 1.0 | 4.1 | GO:0042247 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 1.0 | 6.8 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.9 | 16.1 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.9 | 3.8 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.9 | 4.7 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.9 | 12.2 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.9 | 13.1 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) |
| 0.9 | 16.3 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.9 | 4.4 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) glossopharyngeal nerve development(GO:0021563) |
| 0.9 | 4.4 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.8 | 2.5 | GO:0072334 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.8 | 5.7 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.8 | 2.4 | GO:0002339 | B cell selection(GO:0002339) |
| 0.8 | 7.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.8 | 11.6 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.8 | 3.0 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.7 | 5.8 | GO:0030091 | protein repair(GO:0030091) |
| 0.7 | 10.2 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.7 | 2.1 | GO:0003420 | regulation of growth plate cartilage chondrocyte proliferation(GO:0003420) |
| 0.7 | 4.8 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.7 | 6.7 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.7 | 15.7 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.6 | 5.2 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.6 | 5.0 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.6 | 6.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.6 | 6.7 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.6 | 15.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.6 | 3.0 | GO:2000674 | regulation of type B pancreatic cell apoptotic process(GO:2000674) |
| 0.6 | 7.7 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.6 | 2.9 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.6 | 5.8 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.6 | 4.0 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.6 | 7.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.6 | 1.7 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.5 | 4.3 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.5 | 2.6 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.5 | 5.6 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.5 | 1.5 | GO:0060974 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) |
| 0.5 | 5.4 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.5 | 9.1 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.5 | 6.6 | GO:1903441 | protein localization to ciliary membrane(GO:1903441) |
| 0.5 | 1.4 | GO:1903181 | positive regulation of Wnt protein secretion(GO:0061357) regulation of late endosome to lysosome transport(GO:1902822) regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) negative regulation of vacuolar transport(GO:1903336) |
| 0.4 | 4.8 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.4 | 6.0 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.4 | 7.8 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.4 | 8.8 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.4 | 14.3 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.4 | 4.0 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.4 | 3.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.4 | 16.2 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.4 | 3.9 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.4 | 4.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.4 | 16.8 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.4 | 11.8 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.4 | 5.8 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.3 | 3.4 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.3 | 13.1 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.3 | 17.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.3 | 7.3 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.3 | 3.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.3 | 24.0 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.3 | 2.3 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.3 | 4.6 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.3 | 10.8 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.3 | 20.1 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.3 | 4.3 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.3 | 2.1 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.3 | 1.7 | GO:1990034 | calcium ion export(GO:1901660) calcium ion export from cell(GO:1990034) |
| 0.3 | 0.8 | GO:0036484 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.3 | 9.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.3 | 5.4 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.3 | 0.8 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.3 | 9.2 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.3 | 8.2 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.3 | 6.7 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.2 | 5.7 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.2 | 1.9 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.2 | 3.4 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.2 | 11.0 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.2 | 1.3 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.2 | 3.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.2 | 4.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 3.0 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.2 | 2.1 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.2 | 12.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.2 | 15.3 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.2 | 1.2 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.2 | 13.6 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.2 | 0.5 | GO:0070602 | regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.2 | 5.0 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.2 | 1.7 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.2 | 5.6 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.2 | 2.6 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.2 | 0.8 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.2 | 1.5 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.2 | 0.6 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.2 | 8.0 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.2 | 2.2 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.2 | 12.2 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.2 | 3.0 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.2 | 0.8 | GO:0021764 | amygdala development(GO:0021764) |
| 0.1 | 3.7 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.1 | 8.3 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.1 | 5.9 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.1 | 1.9 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.1 | 3.6 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.1 | 3.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 1.2 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.1 | 0.4 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.1 | 0.9 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 4.9 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 3.9 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.1 | 1.8 | GO:2000188 | regulation of cholesterol homeostasis(GO:2000188) |
| 0.1 | 1.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 8.3 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.1 | 10.3 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.1 | 0.5 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.1 | 0.7 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.1 | 9.1 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 | 0.6 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 6.3 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 6.8 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.1 | 3.2 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 0.8 | GO:0031034 | myosin filament organization(GO:0031033) myosin filament assembly(GO:0031034) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 14.1 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.1 | 0.3 | GO:0071504 | response to heparin(GO:0071503) cellular response to heparin(GO:0071504) |
| 0.1 | 12.1 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.0 | 0.7 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 1.6 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 2.3 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 1.1 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 12.4 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 1.4 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.3 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.0 | 1.6 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 1.6 | GO:0008593 | regulation of Notch signaling pathway(GO:0008593) |
| 0.0 | 3.4 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.0 | 1.8 | GO:0030516 | regulation of axon extension(GO:0030516) |
| 0.0 | 0.2 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.3 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.0 | 0.6 | GO:0009712 | catecholamine metabolic process(GO:0006584) catechol-containing compound metabolic process(GO:0009712) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.3 | 65.3 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 5.0 | 20.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 3.9 | 31.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 3.5 | 21.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 3.3 | 19.6 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 3.0 | 47.7 | GO:0005642 | annulate lamellae(GO:0005642) |
| 2.7 | 10.9 | GO:0071942 | XPC complex(GO:0071942) |
| 2.3 | 27.9 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 2.3 | 6.9 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 2.1 | 10.7 | GO:0097513 | myosin II filament(GO:0097513) |
| 2.1 | 6.3 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 2.0 | 10.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 1.8 | 19.9 | GO:0005638 | lamin filament(GO:0005638) |
| 1.8 | 10.8 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 1.7 | 38.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 1.7 | 6.6 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 1.7 | 9.9 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 1.6 | 9.7 | GO:1990357 | terminal web(GO:1990357) |
| 1.3 | 16.1 | GO:0090543 | Flemming body(GO:0090543) |
| 1.2 | 12.9 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 1.2 | 34.8 | GO:0030057 | desmosome(GO:0030057) |
| 1.1 | 8.7 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 1.1 | 5.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 1.0 | 60.9 | GO:0031430 | M band(GO:0031430) |
| 1.0 | 6.8 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.9 | 12.3 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.9 | 12.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.9 | 17.4 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.9 | 6.0 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.9 | 14.5 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.8 | 4.1 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.8 | 10.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.7 | 13.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.7 | 27.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.7 | 5.6 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.7 | 14.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.6 | 12.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.6 | 5.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.6 | 13.0 | GO:0046930 | pore complex(GO:0046930) |
| 0.6 | 6.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.5 | 36.7 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.5 | 3.6 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.5 | 6.0 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.5 | 1.4 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.4 | 4.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.4 | 4.0 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.4 | 2.6 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.4 | 3.0 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.4 | 6.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.4 | 14.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.4 | 38.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.4 | 3.9 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.4 | 8.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.3 | 3.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.3 | 6.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.3 | 3.0 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.3 | 28.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.3 | 7.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.3 | 1.0 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.3 | 5.0 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.3 | 5.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 21.4 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.2 | 7.8 | GO:0043218 | compact myelin(GO:0043218) |
| 0.2 | 1.7 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.2 | 1.9 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.2 | 5.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 25.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 3.4 | GO:0043219 | lateral loop(GO:0043219) |
| 0.2 | 6.6 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.2 | 4.0 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 33.0 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.2 | 3.5 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.2 | 12.9 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 9.9 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.2 | 6.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 1.6 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 0.7 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 56.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.2 | 1.9 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.2 | 1.4 | GO:0000938 | GARP complex(GO:0000938) |
| 0.2 | 4.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 4.4 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 4.5 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.6 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 13.1 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 10.8 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 18.5 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 10.5 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 1.7 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 11.7 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.1 | 8.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 4.2 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 13.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 3.4 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.1 | 3.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 4.5 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 6.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 0.3 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.1 | 2.2 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 0.9 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.7 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.1 | 6.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 8.5 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 0.7 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 2.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 5.9 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 1.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 6.3 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 4.9 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 10.4 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 3.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 80.5 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.1 | 6.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 4.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 20.8 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 84.6 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 1.1 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 1.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.0 | 84.9 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 15.2 | 45.7 | GO:0048030 | disaccharide binding(GO:0048030) |
| 8.0 | 80.4 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 7.3 | 21.9 | GO:0003974 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 5.5 | 60.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 4.8 | 19.2 | GO:0004420 | hydroxymethylglutaryl-CoA reductase (NADPH) activity(GO:0004420) hydroxymethylglutaryl-CoA reductase activity(GO:0042282) |
| 4.0 | 12.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 3.6 | 28.8 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 3.4 | 20.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 3.3 | 23.3 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 3.1 | 21.6 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 2.9 | 11.8 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 2.6 | 7.7 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 2.5 | 10.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 2.3 | 16.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 2.0 | 6.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 2.0 | 24.0 | GO:0038132 | neuregulin binding(GO:0038132) |
| 1.9 | 45.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 1.7 | 18.2 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 1.6 | 11.5 | GO:0000295 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) ATP transmembrane transporter activity(GO:0005347) purine nucleotide transmembrane transporter activity(GO:0015216) ADP transmembrane transporter activity(GO:0015217) |
| 1.6 | 24.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 1.6 | 4.8 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 1.6 | 7.9 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 1.4 | 11.6 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 1.4 | 28.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 1.4 | 9.9 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 1.3 | 10.6 | GO:0015288 | porin activity(GO:0015288) |
| 1.3 | 3.9 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 1.2 | 11.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 1.2 | 13.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 1.2 | 7.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 1.2 | 4.7 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 1.2 | 5.8 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 1.1 | 9.1 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 1.1 | 4.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 1.0 | 7.8 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 1.0 | 3.9 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.9 | 17.1 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.9 | 9.9 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.9 | 4.4 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.9 | 9.4 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.8 | 2.5 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.8 | 4.8 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.8 | 24.3 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.8 | 46.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.7 | 6.5 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.7 | 5.0 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.7 | 6.7 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.7 | 3.3 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.6 | 17.2 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.6 | 3.0 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.6 | 28.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.6 | 8.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.5 | 12.3 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.5 | 6.9 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.5 | 9.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.5 | 5.3 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.5 | 9.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.5 | 3.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.5 | 6.0 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.5 | 10.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.5 | 6.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.4 | 11.6 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.4 | 3.3 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.4 | 26.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.4 | 14.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.4 | 12.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.4 | 11.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.4 | 1.5 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.3 | 8.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.3 | 1.7 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.3 | 2.6 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.3 | 15.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.3 | 2.5 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.3 | 12.1 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.3 | 69.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.3 | 5.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.3 | 3.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.3 | 2.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.3 | 13.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.3 | 9.9 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.2 | 1.0 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.2 | 8.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 11.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.2 | 3.7 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 5.7 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.2 | 12.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 2.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.2 | 6.6 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.2 | 5.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 32.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.2 | 7.9 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.2 | 0.5 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.2 | 15.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.2 | 19.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 3.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 3.8 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.2 | 0.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 0.6 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 1.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 2.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 2.3 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 49.0 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 9.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.8 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 13.5 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 9.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.3 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.1 | 2.6 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 7.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 2.4 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 1.7 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.1 | 1.1 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.1 | 1.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 3.5 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 0.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 19.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 0.3 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 3.4 | GO:0004407 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.1 | 0.6 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 5.4 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 6.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 2.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 1.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 13.6 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 5.5 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 2.9 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 1.4 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 0.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 2.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.8 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.2 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 18.1 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 4.4 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 3.1 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.7 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 1.0 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 0.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 23.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 1.1 | 47.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 1.1 | 22.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 1.1 | 22.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 1.1 | 20.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.9 | 6.6 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.8 | 19.5 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.8 | 151.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.8 | 3.0 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.7 | 9.6 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.6 | 10.4 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.5 | 39.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.5 | 2.6 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.5 | 58.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.4 | 14.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.4 | 13.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.4 | 29.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.4 | 22.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.3 | 17.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.3 | 13.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.3 | 18.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.3 | 14.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.3 | 20.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.3 | 30.4 | PID P73PATHWAY | p73 transcription factor network |
| 0.3 | 19.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 4.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 20.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 9.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 4.8 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.2 | 20.2 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.2 | 6.9 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.2 | 17.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.2 | 6.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 4.8 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 5.2 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 4.7 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.1 | 7.3 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 2.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 2.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 3.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 5.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 5.1 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 2.6 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 2.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.1 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 5.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 9.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 20.6 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 1.4 | 41.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 1.3 | 74.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 1.1 | 48.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.9 | 22.0 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.9 | 19.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.8 | 30.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.7 | 19.5 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.7 | 4.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.7 | 32.8 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.7 | 21.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.7 | 18.2 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.7 | 11.8 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.7 | 4.8 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.7 | 28.2 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.6 | 16.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.6 | 21.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.6 | 18.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.5 | 4.7 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.4 | 10.9 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.4 | 10.0 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.4 | 19.9 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.4 | 14.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.4 | 11.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.4 | 9.8 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.3 | 10.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.3 | 11.0 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.3 | 9.9 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.3 | 5.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.3 | 0.9 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.3 | 6.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.3 | 37.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.3 | 13.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.3 | 20.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 8.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 4.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 5.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 8.7 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.2 | 1.9 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.2 | 14.9 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.2 | 5.4 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.2 | 12.6 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.2 | 16.9 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.2 | 3.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 2.9 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 3.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 0.8 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 5.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 2.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 3.0 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 7.9 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 2.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 0.7 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 5.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 19.9 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.1 | 3.4 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 2.4 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 11.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 2.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.5 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 4.6 | REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | Genes involved in MyD88:Mal cascade initiated on plasma membrane |
| 0.0 | 2.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.8 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 1.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 3.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 8.0 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 2.9 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |