Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
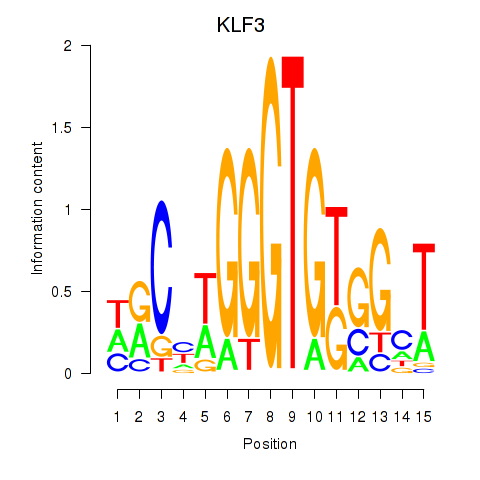
Results for KLF3
Z-value: 1.05
Transcription factors associated with KLF3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF3
|
ENSG00000109787.13 | KLF3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF3 | hg38_v1_chr4_+_38664189_38664206 | 0.37 | 2.8e-08 | Click! |
Activity profile of KLF3 motif
Sorted Z-values of KLF3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 23.9 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 3.5 | 20.7 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 3.4 | 10.2 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 2.9 | 11.7 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 2.6 | 7.9 | GO:1902490 | regulation of sperm capacitation(GO:1902490) |
| 2.6 | 44.8 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 2.5 | 15.2 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 2.2 | 17.9 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 2.1 | 8.5 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 2.1 | 27.4 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 2.0 | 2.0 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 2.0 | 6.1 | GO:0051939 | gamma-aminobutyric acid import(GO:0051939) |
| 2.0 | 6.1 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 1.9 | 7.7 | GO:0071879 | UDP-glucose catabolic process(GO:0006258) positive regulation of adrenergic receptor signaling pathway(GO:0071879) negative regulation of type B pancreatic cell development(GO:2000077) |
| 1.6 | 4.9 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 1.6 | 6.5 | GO:1904021 | regulation of G-protein coupled receptor internalization(GO:1904020) negative regulation of G-protein coupled receptor internalization(GO:1904021) |
| 1.5 | 4.4 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 1.5 | 7.4 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 1.4 | 4.3 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) beta-alanine metabolic process(GO:0019482) thymine metabolic process(GO:0019859) |
| 1.4 | 8.3 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 1.4 | 6.8 | GO:1901523 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 1.3 | 4.0 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 1.3 | 5.2 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 1.3 | 5.0 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 1.2 | 18.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.1 | 5.6 | GO:0051029 | rRNA transport(GO:0051029) |
| 1.1 | 3.3 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 1.1 | 8.8 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 1.1 | 3.2 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 1.0 | 4.2 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 1.0 | 4.1 | GO:0002215 | defense response to nematode(GO:0002215) |
| 1.0 | 3.0 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 1.0 | 8.7 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 1.0 | 8.7 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 1.0 | 3.8 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.9 | 2.8 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.9 | 2.7 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.9 | 2.6 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.9 | 2.6 | GO:1902771 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.8 | 8.3 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.8 | 2.4 | GO:2000359 | regulation of binding of sperm to zona pellucida(GO:2000359) negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.8 | 4.0 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.8 | 15.8 | GO:0048757 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.8 | 10.0 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.8 | 8.3 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.7 | 2.9 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.7 | 6.6 | GO:0070054 | mRNA splicing via endonucleolytic cleavage and ligation involved in unfolded protein response(GO:0030969) mRNA splicing, via endonucleolytic cleavage and ligation(GO:0070054) mRNA endonucleolytic cleavage involved in unfolded protein response(GO:0070055) |
| 0.7 | 4.4 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.7 | 2.9 | GO:1902617 | response to fluoride(GO:1902617) |
| 0.7 | 12.8 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.7 | 2.1 | GO:0070408 | carbamoyl phosphate metabolic process(GO:0070408) carbamoyl phosphate biosynthetic process(GO:0070409) cellular response to oleic acid(GO:0071400) response to ammonia(GO:1903717) cellular response to ammonia(GO:1903718) |
| 0.7 | 2.1 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.7 | 2.0 | GO:0048627 | myoblast development(GO:0048627) |
| 0.7 | 2.0 | GO:1903238 | positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.7 | 6.0 | GO:1901731 | positive regulation of platelet aggregation(GO:1901731) |
| 0.7 | 1.3 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.7 | 6.5 | GO:0090160 | endosomal vesicle fusion(GO:0034058) Golgi to lysosome transport(GO:0090160) |
| 0.6 | 1.9 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.6 | 5.2 | GO:0086036 | regulation of cardiac muscle cell membrane potential(GO:0086036) |
| 0.6 | 4.5 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.6 | 3.5 | GO:0006050 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.6 | 1.7 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.6 | 4.6 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.6 | 1.7 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) |
| 0.6 | 12.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.6 | 1.1 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.5 | 1.6 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.5 | 1.6 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.5 | 2.1 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.5 | 4.8 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.5 | 3.2 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.5 | 4.7 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.5 | 1.5 | GO:0060032 | notochord regression(GO:0060032) |
| 0.5 | 4.0 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.5 | 5.4 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.5 | 3.4 | GO:1903236 | regulation of leukocyte tethering or rolling(GO:1903236) negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.5 | 2.9 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.5 | 4.6 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.5 | 1.8 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.4 | 1.8 | GO:0009137 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.4 | 1.7 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.4 | 3.0 | GO:0072103 | glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.4 | 2.9 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.4 | 1.7 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.4 | 4.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.4 | 6.8 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.4 | 1.2 | GO:0070843 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.4 | 1.6 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.4 | 1.2 | GO:0086047 | membrane depolarization during Purkinje myocyte cell action potential(GO:0086047) |
| 0.4 | 1.5 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.4 | 6.4 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.4 | 1.1 | GO:0043605 | antibiotic metabolic process(GO:0016999) cellular amide catabolic process(GO:0043605) |
| 0.4 | 1.5 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.4 | 1.1 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.4 | 5.8 | GO:0070444 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.4 | 2.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.4 | 3.9 | GO:1904152 | regulation of retrograde protein transport, ER to cytosol(GO:1904152) |
| 0.3 | 5.5 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.3 | 1.7 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.3 | 4.4 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.3 | 3.0 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.3 | 6.1 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.3 | 3.5 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.3 | 0.9 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.3 | 1.5 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.3 | 0.3 | GO:0038129 | ERBB3 signaling pathway(GO:0038129) |
| 0.3 | 2.9 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.3 | 1.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.3 | 2.8 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.3 | 7.4 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.3 | 3.7 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.3 | 1.9 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.3 | 0.8 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.3 | 5.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.3 | 2.7 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.3 | 7.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.3 | 1.6 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.2 | 4.4 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.2 | 0.7 | GO:0042441 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.2 | 8.7 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.2 | 2.8 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.2 | 1.6 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.2 | 2.3 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.2 | 14.7 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.2 | 0.9 | GO:0014806 | smooth muscle hyperplasia(GO:0014806) negative regulation of mast cell cytokine production(GO:0032764) |
| 0.2 | 0.7 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.2 | 4.4 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.2 | 1.5 | GO:0036484 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.2 | 2.6 | GO:0035878 | nail development(GO:0035878) |
| 0.2 | 2.9 | GO:0060452 | positive regulation of cardiac muscle contraction(GO:0060452) |
| 0.2 | 1.8 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.2 | 0.6 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.2 | 2.1 | GO:0050665 | hydrogen peroxide biosynthetic process(GO:0050665) |
| 0.2 | 2.7 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.2 | 1.9 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.2 | 3.8 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.2 | 1.7 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.2 | 2.5 | GO:0046520 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.2 | 3.0 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.2 | 1.8 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.2 | 1.9 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.2 | 2.8 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.2 | 1.2 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.2 | 2.6 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.2 | 2.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 2.5 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.2 | 3.9 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.2 | 0.8 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 0.2 | 1.7 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.2 | 2.8 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.2 | 2.0 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.2 | 0.5 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.2 | 1.9 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.1 | 8.5 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 1.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 0.4 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 6.6 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 4.0 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 2.5 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 4.0 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.1 | 2.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 4.4 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.1 | 4.3 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.1 | 3.5 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 2.0 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.4 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.1 | 0.8 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.1 | 0.9 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.1 | 1.8 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 1.0 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 2.0 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 0.6 | GO:0036018 | cellular response to erythropoietin(GO:0036018) positive regulation of protein kinase C signaling(GO:0090037) |
| 0.1 | 1.1 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 1.0 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 0.9 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.1 | 2.9 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.3 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.1 | 0.7 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.1 | 10.8 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 2.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.6 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 2.2 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 0.4 | GO:0086029 | Purkinje myocyte to ventricular cardiac muscle cell signaling(GO:0086029) Purkinje myocyte to ventricular cardiac muscle cell communication(GO:0086068) |
| 0.1 | 1.2 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.6 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 2.7 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 4.4 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 0.6 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.9 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 3.0 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 0.6 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 | 0.8 | GO:0045348 | positive regulation of MHC class I biosynthetic process(GO:0045345) positive regulation of MHC class II biosynthetic process(GO:0045348) |
| 0.1 | 4.3 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.1 | 3.6 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.1 | 2.4 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.1 | 2.1 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.1 | 5.1 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 2.7 | GO:0043304 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.1 | 1.5 | GO:0061436 | regulation of water loss via skin(GO:0033561) establishment of skin barrier(GO:0061436) |
| 0.1 | 3.4 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.1 | 1.9 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.1 | 0.4 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 1.6 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 1.3 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.1 | 5.4 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) |
| 0.1 | 2.4 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 1.0 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.1 | 2.6 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.1 | 0.5 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 0.6 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.1 | 2.4 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.1 | 0.7 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 2.0 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 4.7 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 | 1.0 | GO:0007620 | copulation(GO:0007620) |
| 0.1 | 2.5 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 3.4 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 1.0 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 0.5 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.1 | 0.4 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.4 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.1 | 0.9 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 2.2 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 9.0 | GO:0007565 | female pregnancy(GO:0007565) |
| 0.0 | 0.9 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 8.7 | GO:0030518 | intracellular steroid hormone receptor signaling pathway(GO:0030518) |
| 0.0 | 0.9 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.4 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 3.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 2.6 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.4 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 | 0.7 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.6 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.7 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 3.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 3.9 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 2.3 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.7 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.0 | 2.0 | GO:0032729 | positive regulation of interferon-gamma production(GO:0032729) |
| 0.0 | 0.8 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 1.2 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 4.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.2 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 1.3 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 1.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.2 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.3 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 1.5 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 2.7 | GO:0007612 | learning(GO:0007612) |
| 0.0 | 4.6 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 1.2 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 0.0 | GO:0003418 | growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 13.3 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 1.0 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.7 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 1.6 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 0.5 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 1.5 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.4 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 1.4 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 2.1 | GO:0050890 | cognition(GO:0050890) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.3 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.0 | 0.7 | GO:0050909 | sensory perception of taste(GO:0050909) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 23.9 | GO:0045179 | apical cortex(GO:0045179) |
| 2.2 | 6.6 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 1.6 | 14.7 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 1.4 | 5.6 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 1.3 | 5.2 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 1.2 | 5.8 | GO:0072534 | perineuronal net(GO:0072534) |
| 1.1 | 20.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 1.0 | 2.9 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.9 | 18.5 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.9 | 9.0 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.9 | 2.6 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.8 | 2.4 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.7 | 4.2 | GO:0002177 | manchette(GO:0002177) |
| 0.7 | 5.2 | GO:0070187 | telosome(GO:0070187) |
| 0.6 | 4.9 | GO:0060199 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.6 | 5.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.5 | 8.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.5 | 1.9 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.4 | 7.9 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.4 | 1.7 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.4 | 6.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.4 | 11.7 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.4 | 2.8 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.4 | 2.4 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.4 | 1.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.4 | 1.9 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.4 | 7.7 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.3 | 4.9 | GO:0033179 | proton-transporting two-sector ATPase complex, proton-transporting domain(GO:0033177) proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.3 | 2.4 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.3 | 8.5 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.3 | 2.9 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.3 | 7.4 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.3 | 4.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.2 | 7.4 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.2 | 2.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 6.0 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 2.9 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 1.8 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.2 | 4.2 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.2 | 15.2 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.2 | 42.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.2 | 2.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.2 | 4.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.2 | 2.0 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 3.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 1.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 1.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 2.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.2 | 3.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 2.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.2 | 2.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 7.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 1.4 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 3.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 0.9 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 0.8 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 2.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 4.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 1.0 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 4.0 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 0.7 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 3.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 11.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.9 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.1 | 3.0 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 0.6 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 4.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 2.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 4.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 1.7 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 2.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 1.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 11.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 1.2 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 7.8 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 1.6 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 8.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 3.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 6.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 1.1 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 5.3 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 8.1 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 3.9 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 2.6 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 0.6 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 1.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.7 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 2.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 2.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 4.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 2.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 6.7 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 2.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 2.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 2.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 2.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 9.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 2.8 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 2.2 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 1.4 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 0.7 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 3.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 7.8 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 9.0 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 1.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 3.3 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 1.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 5.6 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 9.4 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.1 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.4 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 2.9 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 12.8 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 2.9 | 11.7 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 2.2 | 8.8 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 2.2 | 6.5 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 1.8 | 5.4 | GO:0052870 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 1.7 | 5.0 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 1.7 | 5.0 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 1.6 | 19.0 | GO:0008430 | selenium binding(GO:0008430) |
| 1.5 | 4.6 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 1.5 | 7.4 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 1.4 | 7.2 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 1.4 | 4.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 1.4 | 18.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.3 | 10.5 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 1.3 | 5.2 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 1.3 | 20.7 | GO:0050693 | LBD domain binding(GO:0050693) |
| 1.2 | 8.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 1.2 | 1.2 | GO:0086062 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 1.1 | 3.4 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 1.0 | 23.9 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 1.0 | 4.0 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 1.0 | 4.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 1.0 | 4.9 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 1.0 | 3.9 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.9 | 2.7 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.9 | 5.4 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.9 | 3.6 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) |
| 0.8 | 4.0 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.7 | 6.7 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.7 | 2.2 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.7 | 2.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.7 | 3.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.7 | 4.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.7 | 3.3 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.7 | 8.7 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.7 | 2.0 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.7 | 2.0 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.7 | 15.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.6 | 5.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.6 | 2.6 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.6 | 2.4 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.6 | 1.7 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.6 | 3.4 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.6 | 1.7 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.6 | 2.8 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.5 | 1.6 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.5 | 1.6 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.5 | 2.1 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.5 | 1.6 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.5 | 14.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.5 | 2.0 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.4 | 2.7 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.4 | 1.8 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.4 | 7.9 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.4 | 11.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.4 | 2.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.4 | 1.7 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.4 | 2.9 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.4 | 1.2 | GO:0047783 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.4 | 5.2 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.4 | 4.0 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.4 | 2.0 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.4 | 6.6 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.4 | 3.0 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.4 | 3.0 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.4 | 1.1 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.4 | 2.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.3 | 4.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.3 | 10.6 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.3 | 4.0 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.3 | 2.9 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.3 | 8.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.3 | 3.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.3 | 1.5 | GO:1901474 | L-alanine transmembrane transporter activity(GO:0015180) L-aspartate transmembrane transporter activity(GO:0015183) alanine transmembrane transporter activity(GO:0022858) azole transmembrane transporter activity(GO:1901474) |
| 0.3 | 0.9 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.3 | 1.5 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.3 | 3.0 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.3 | 1.2 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.3 | 2.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.3 | 1.1 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.3 | 8.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.3 | 2.7 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.3 | 7.0 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.3 | 1.9 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.3 | 12.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.3 | 6.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.3 | 10.0 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.3 | 16.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.3 | 1.0 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.3 | 8.3 | GO:0001848 | complement binding(GO:0001848) |
| 0.3 | 7.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 7.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 2.7 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.2 | 3.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 1.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 1.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.2 | 2.4 | GO:0008061 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.2 | 5.6 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.2 | 0.4 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.2 | 3.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 2.7 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.2 | 2.9 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.2 | 1.2 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.2 | 2.7 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.2 | 0.8 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.2 | 1.0 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.2 | 2.7 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 8.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.2 | 5.8 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 0.9 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) phospholipase D activity(GO:0004630) |
| 0.2 | 2.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.2 | 1.8 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.2 | 8.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.2 | 9.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 3.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.2 | 1.0 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.2 | 3.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 6.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.2 | 1.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 2.8 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 2.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.2 | 2.3 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 15.7 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 0.6 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 1.9 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 1.9 | GO:0008179 | guanylate cyclase activity(GO:0004383) adenylate cyclase binding(GO:0008179) |
| 0.1 | 1.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 5.2 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 0.6 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.1 | 2.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 1.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 2.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 1.8 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 0.7 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 3.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.6 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 6.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.6 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 2.9 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 0.8 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 2.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 1.0 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 21.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 2.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 1.5 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.1 | 2.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 7.8 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 3.4 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 1.0 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 5.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 3.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 2.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.3 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 1.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 1.9 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 2.0 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.7 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 3.0 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 5.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 3.8 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 0.8 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 3.4 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 3.1 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.7 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.7 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 4.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 14.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 2.1 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.0 | 3.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.2 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 2.2 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.5 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 2.3 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 1.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 3.1 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 0.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 1.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.4 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 2.6 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.0 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.6 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 1.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 2.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.9 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 44.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.4 | 0.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 48.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.3 | 10.6 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.3 | 5.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.3 | 3.2 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.2 | 2.6 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 3.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.2 | 1.0 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.2 | 2.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 3.3 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.2 | 4.8 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.2 | 18.9 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.2 | 16.4 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.2 | 11.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 4.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 9.7 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 2.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 3.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 19.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 3.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 1.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 3.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 4.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 5.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 18.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.4 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 2.0 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 2.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 2.6 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 0.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 0.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 2.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 2.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 2.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 2.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.0 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.7 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 2.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 3.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 3.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.1 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 2.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 2.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.1 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.6 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.3 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 1.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 7.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 2.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 7.2 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 1.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 15.8 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.9 | 19.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.8 | 12.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.8 | 15.0 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.7 | 3.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.7 | 18.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.7 | 5.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.4 | 9.8 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.4 | 3.0 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.4 | 11.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.4 | 11.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.3 | 8.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 10.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.3 | 5.6 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.3 | 3.8 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.3 | 7.0 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.3 | 4.5 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.2 | 3.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.2 | 9.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 10.1 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.2 | 2.9 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 12.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.2 | 4.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 6.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.2 | 10.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 2.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 5.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.2 | 2.7 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 1.9 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 2.9 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.1 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.1 | 6.8 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 2.1 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 3.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 3.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.7 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 2.0 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 3.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 3.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 2.7 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 2.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 4.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 2.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 3.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.7 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 6.4 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 5.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 11.4 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 3.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 4.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.0 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.8 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 5.0 | REACTOME UNFOLDED PROTEIN RESPONSE | Genes involved in Unfolded Protein Response |
| 0.0 | 0.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 3.8 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 3.8 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 15.2 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.4 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 1.9 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 1.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.2 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.1 | REACTOME PI3K EVENTS IN ERBB4 SIGNALING | Genes involved in PI3K events in ERBB4 signaling |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.0 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |