Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
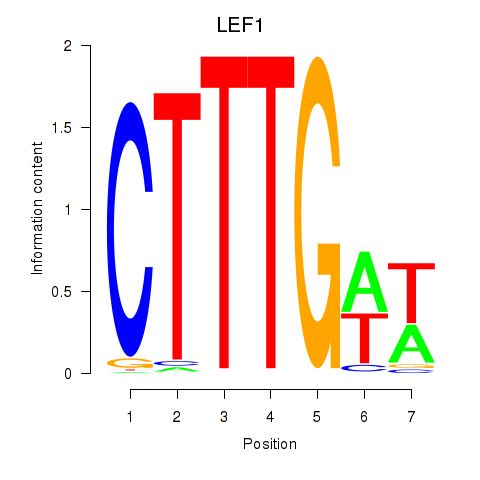
Results for LEF1
Z-value: 0.90
Transcription factors associated with LEF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LEF1
|
ENSG00000138795.10 | LEF1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LEF1 | hg38_v1_chr4_-_108168919_108168939 | -0.07 | 2.7e-01 | Click! |
Activity profile of LEF1 motif
Sorted Z-values of LEF1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of LEF1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 20.9 | GO:0032599 | B cell receptor transport within lipid bilayer(GO:0032595) B cell receptor transport into membrane raft(GO:0032597) protein transport out of membrane raft(GO:0032599) chemokine receptor transport out of membrane raft(GO:0032600) negative regulation of transforming growth factor beta3 production(GO:0032913) chemokine receptor transport within lipid bilayer(GO:0033606) |
| 4.9 | 14.6 | GO:0009996 | negative regulation of cell fate specification(GO:0009996) |
| 4.0 | 16.0 | GO:0002188 | translation reinitiation(GO:0002188) |
| 3.8 | 15.3 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 3.2 | 12.7 | GO:0009956 | radial pattern formation(GO:0009956) |
| 3.0 | 9.1 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 2.9 | 8.7 | GO:0051821 | negative regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010768) transmission of virus(GO:0019089) dissemination or transmission of symbiont from host(GO:0044007) dissemination or transmission of organism from other organism involved in symbiotic interaction(GO:0051821) |
| 2.9 | 8.6 | GO:1904899 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 2.7 | 24.5 | GO:0007070 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 2.7 | 13.6 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 2.5 | 7.4 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 2.1 | 6.4 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 2.0 | 8.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 1.7 | 12.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 1.7 | 8.6 | GO:0030035 | microspike assembly(GO:0030035) |
| 1.6 | 27.3 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 1.4 | 5.7 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 1.4 | 6.9 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 1.4 | 2.7 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 1.4 | 5.4 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 1.3 | 12.0 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 1.2 | 10.0 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 1.2 | 3.7 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 1.2 | 6.0 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 1.2 | 3.5 | GO:0048817 | negative regulation of transforming growth factor beta2 production(GO:0032912) negative regulation of hair follicle maturation(GO:0048817) regulation of melanosome transport(GO:1902908) |
| 1.1 | 7.5 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 1.0 | 6.2 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 1.0 | 6.2 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 1.0 | 7.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 1.0 | 10.8 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 1.0 | 4.9 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 1.0 | 6.8 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 1.0 | 3.8 | GO:0003290 | atrial septum secundum morphogenesis(GO:0003290) |
| 0.9 | 3.8 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.9 | 6.5 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.9 | 12.9 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.9 | 8.0 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.9 | 5.2 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.9 | 3.4 | GO:1904020 | negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of G-protein coupled receptor internalization(GO:1904020) negative regulation of G-protein coupled receptor internalization(GO:1904021) |
| 0.8 | 2.5 | GO:0097114 | NMDA glutamate receptor clustering(GO:0097114) |
| 0.8 | 2.4 | GO:2000798 | amniotic stem cell differentiation(GO:0097086) negative regulation of dense core granule biogenesis(GO:2000706) negative regulation of mesenchymal stem cell differentiation(GO:2000740) regulation of amniotic stem cell differentiation(GO:2000797) negative regulation of amniotic stem cell differentiation(GO:2000798) |
| 0.8 | 10.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.8 | 2.4 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.8 | 6.8 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.7 | 3.0 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.7 | 2.9 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 0.7 | 4.8 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.7 | 26.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.7 | 4.6 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.6 | 11.0 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.6 | 5.7 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.6 | 2.5 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.6 | 7.8 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.6 | 7.0 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.6 | 5.3 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.6 | 2.3 | GO:0009597 | detection of virus(GO:0009597) |
| 0.6 | 2.9 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.6 | 6.9 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.6 | 2.8 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.6 | 22.8 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.6 | 5.7 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.5 | 1.6 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.5 | 1.6 | GO:0021593 | rhombomere morphogenesis(GO:0021593) rhombomere 3 morphogenesis(GO:0021658) |
| 0.5 | 12.9 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.5 | 2.7 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.5 | 9.6 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.5 | 2.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.5 | 4.3 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.5 | 2.8 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.5 | 48.0 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.5 | 2.7 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.4 | 10.9 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.4 | 4.6 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.4 | 8.5 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.4 | 1.2 | GO:2000176 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.4 | 4.8 | GO:1901631 | positive regulation of presynaptic membrane organization(GO:1901631) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.4 | 5.5 | GO:1904871 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.4 | 2.3 | GO:0035549 | positive regulation of interferon-beta secretion(GO:0035549) |
| 0.4 | 2.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.4 | 17.2 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.4 | 4.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.4 | 4.4 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.3 | 12.6 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.3 | 2.0 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.3 | 3.7 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.3 | 7.7 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.3 | 2.3 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.3 | 5.5 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.3 | 2.6 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.3 | 4.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.3 | 2.8 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.3 | 1.6 | GO:2000317 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.3 | 7.9 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.3 | 1.5 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.3 | 2.3 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.3 | 0.9 | GO:0005988 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.3 | 5.8 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.3 | 4.3 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.3 | 0.9 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.3 | 1.4 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.3 | 2.0 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.3 | 5.4 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.3 | 9.5 | GO:2000810 | regulation of bicellular tight junction assembly(GO:2000810) |
| 0.3 | 1.0 | GO:1904782 | negative regulation of glutamate receptor signaling pathway(GO:1900450) negative regulation of NMDA glutamate receptor activity(GO:1904782) |
| 0.3 | 0.8 | GO:0060003 | copper ion export(GO:0060003) |
| 0.2 | 1.7 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.2 | 1.2 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.2 | 5.9 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.2 | 2.1 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.2 | 5.0 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.2 | 6.4 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.2 | 3.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 1.4 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.2 | 1.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 | 1.5 | GO:1905068 | arterial endothelial cell fate commitment(GO:0060844) blood vessel lumenization(GO:0072554) positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.2 | 0.4 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.2 | 1.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.2 | 1.5 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.2 | 2.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.2 | 3.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 | 0.6 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.2 | 6.8 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.2 | 1.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.2 | 1.7 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.2 | 0.7 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.2 | 2.3 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.2 | 2.0 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.2 | 0.8 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.2 | 0.5 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.2 | 1.8 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.2 | 3.1 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.2 | 0.6 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.2 | 3.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.2 | 4.9 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 3.1 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 0.9 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 0.4 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 2.1 | GO:0035246 | peptidyl-arginine N-methylation(GO:0035246) |
| 0.1 | 2.9 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.1 | 0.8 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 3.0 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 1.4 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 2.9 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.1 | 1.4 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 4.7 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 3.9 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 10.8 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 0.2 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.1 | 1.7 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 | 0.5 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 3.4 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.1 | 0.7 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.1 | 6.0 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.1 | 0.3 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.1 | 4.1 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.1 | 1.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 0.9 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.3 | GO:0048880 | sensory system development(GO:0048880) |
| 0.1 | 3.9 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.1 | 0.8 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 0.3 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 | 1.8 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 3.2 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 0.4 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 8.6 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.1 | 2.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 7.5 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 0.7 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.1 | 8.8 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 1.5 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 3.1 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 1.0 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.1 | 5.6 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.1 | 1.7 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.1 | 0.8 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 0.3 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 5.7 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 1.2 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 0.6 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 2.9 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.1 | 0.4 | GO:0060368 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.1 | 9.6 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.1 | 2.4 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 3.4 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.1 | 1.4 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 0.9 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.1 | 0.5 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 0.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 2.6 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 1.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.9 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 1.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 1.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 2.4 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 1.0 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 | 1.3 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.6 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 12.9 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 1.1 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 1.5 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 0.6 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.3 | GO:0001705 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.9 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.4 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 3.3 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.3 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.3 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 1.1 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.5 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.2 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 1.0 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 2.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 4.1 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 2.0 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.0 | 0.3 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 3.1 | GO:0034101 | erythrocyte homeostasis(GO:0034101) |
| 0.0 | 0.4 | GO:0006882 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.0 | 0.5 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.7 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.9 | GO:0031016 | pancreas development(GO:0031016) |
| 0.0 | 0.1 | GO:2001197 | positive regulation of extracellular matrix assembly(GO:1901203) regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.8 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 1.2 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 2.7 | GO:0006275 | regulation of DNA replication(GO:0006275) |
| 0.0 | 2.1 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 1.0 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 1.2 | GO:0043279 | response to alkaloid(GO:0043279) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 10.2 | GO:0008623 | CHRAC(GO:0008623) |
| 2.3 | 7.0 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 2.1 | 6.4 | GO:0018444 | translation release factor complex(GO:0018444) |
| 1.8 | 12.9 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 1.3 | 3.9 | GO:0044393 | microspike(GO:0044393) |
| 1.3 | 3.8 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 1.1 | 9.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 1.1 | 3.4 | GO:1903349 | omegasome membrane(GO:1903349) |
| 1.1 | 5.3 | GO:0042025 | host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 1.0 | 8.0 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 1.0 | 3.0 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 1.0 | 33.2 | GO:0016514 | SWI/SNF complex(GO:0016514) npBAF complex(GO:0071564) |
| 1.0 | 2.9 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 1.0 | 9.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.9 | 20.9 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.8 | 5.0 | GO:0005816 | spindle pole body(GO:0005816) |
| 0.8 | 10.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.8 | 2.4 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.8 | 6.8 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.7 | 17.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.7 | 9.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.7 | 4.1 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.7 | 6.6 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.7 | 3.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.6 | 2.5 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.6 | 3.0 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.5 | 12.0 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.5 | 6.5 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.5 | 2.6 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.5 | 6.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.5 | 4.7 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.5 | 2.4 | GO:0016589 | NURF complex(GO:0016589) |
| 0.5 | 2.8 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.4 | 5.2 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.4 | 7.7 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.4 | 3.8 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.4 | 8.0 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.4 | 3.9 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.4 | 5.9 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.4 | 5.5 | GO:0070938 | contractile ring(GO:0070938) |
| 0.4 | 19.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.4 | 2.5 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.3 | 5.5 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.3 | 9.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.3 | 8.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.3 | 19.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.3 | 4.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.3 | 15.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 5.5 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.2 | 0.7 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.2 | 1.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.2 | 2.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 3.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.2 | 2.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 9.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 9.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.2 | 3.7 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.2 | 1.5 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.2 | 24.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 1.9 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 1.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 1.4 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 0.7 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 10.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 16.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 1.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 2.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 0.9 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.1 | 2.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 2.7 | GO:0000791 | euchromatin(GO:0000791) |
| 0.1 | 9.7 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 8.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 2.9 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 5.2 | GO:0043657 | host(GO:0018995) host cell(GO:0043657) |
| 0.1 | 1.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.5 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 32.1 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 4.1 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 2.0 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 1.0 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 0.3 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 7.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 1.0 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.1 | 15.9 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.1 | 9.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 8.4 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 5.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 1.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 4.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.0 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 3.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.2 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 10.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 3.0 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 2.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 2.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.4 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 4.2 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 0.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 2.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.6 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 1.3 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 1.4 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 15.3 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 1.7 | 5.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 1.4 | 15.6 | GO:0039706 | co-receptor binding(GO:0039706) |
| 1.3 | 24.5 | GO:0030957 | Tat protein binding(GO:0030957) |
| 1.2 | 8.7 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 1.2 | 10.0 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 1.1 | 5.5 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 1.0 | 7.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 1.0 | 21.1 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 1.0 | 6.0 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 1.0 | 2.9 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 0.9 | 4.7 | GO:0004515 | nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.9 | 4.6 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.9 | 6.4 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.9 | 16.8 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.9 | 4.4 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.9 | 3.5 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.9 | 5.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.9 | 8.6 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.8 | 4.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.8 | 10.8 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.7 | 3.0 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.7 | 7.4 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.7 | 13.0 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.7 | 2.1 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.7 | 41.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.7 | 8.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.6 | 5.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.6 | 3.7 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.6 | 2.3 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.6 | 12.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.6 | 2.2 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.6 | 15.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.6 | 21.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.5 | 3.8 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.5 | 4.3 | GO:0070697 | activin receptor binding(GO:0070697) type II activin receptor binding(GO:0070699) |
| 0.5 | 6.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.5 | 14.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.5 | 1.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.5 | 2.0 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.5 | 3.0 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.5 | 23.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.5 | 6.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.5 | 5.7 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.5 | 7.0 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.5 | 2.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.4 | 14.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.4 | 2.8 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.4 | 2.0 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.4 | 6.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.4 | 17.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.4 | 1.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.4 | 25.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.4 | 1.8 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.3 | 13.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.3 | 5.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.3 | 6.0 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.3 | 26.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.3 | 1.5 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.3 | 1.5 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.3 | 0.9 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.3 | 2.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.3 | 0.9 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.3 | 7.9 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.3 | 7.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.3 | 0.8 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.3 | 0.8 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.3 | 13.2 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.2 | 2.0 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.2 | 1.2 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.2 | 1.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.2 | 0.7 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.2 | 0.9 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.2 | 6.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 9.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.2 | 1.4 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.2 | 0.4 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.2 | 2.3 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.2 | 2.9 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.2 | 8.3 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.2 | 7.1 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.2 | 1.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.2 | 3.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.2 | 2.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 0.8 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.2 | 11.1 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 4.4 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 2.1 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.1 | 1.6 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 3.9 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 1.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 13.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 0.8 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 2.4 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.1 | 0.5 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 1.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 3.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 1.8 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 2.5 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 1.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 3.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 2.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.5 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 1.5 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 1.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.1 | 4.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.5 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 2.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 8.9 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 1.5 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 38.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 3.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 2.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.3 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 2.4 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.1 | 0.2 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.1 | 2.6 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 1.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 5.6 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 11.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 1.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 35.1 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.1 | 0.8 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 7.1 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 11.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 0.3 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.7 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 7.2 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 16.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.6 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 1.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.2 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.8 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 1.7 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 6.6 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.6 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.9 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 1.8 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 0.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 2.2 | GO:0000287 | magnesium ion binding(GO:0000287) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 24.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.5 | 10.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.5 | 37.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.4 | 8.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.4 | 12.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.4 | 23.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.3 | 13.6 | PID MYC PATHWAY | C-MYC pathway |
| 0.3 | 13.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 28.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.3 | 11.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.3 | 28.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.3 | 16.0 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.3 | 5.8 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.2 | 3.4 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.2 | 7.5 | PID ATR PATHWAY | ATR signaling pathway |
| 0.2 | 8.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.2 | 17.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.2 | 4.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.2 | 5.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 12.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 2.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 5.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.2 | 9.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 19.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.2 | 10.5 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 1.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 9.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 1.0 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 1.2 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 6.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 2.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 2.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 3.6 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 1.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 1.6 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 2.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 2.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 4.6 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 0.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.4 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.4 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 26.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.8 | 15.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.7 | 18.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.7 | 10.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.7 | 12.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.5 | 15.7 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.5 | 23.9 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.5 | 13.8 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.4 | 13.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.4 | 15.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.4 | 13.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.3 | 7.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.3 | 10.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.3 | 0.9 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.3 | 3.4 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.3 | 5.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.3 | 7.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 8.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.3 | 9.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.3 | 2.1 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.3 | 42.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.2 | 3.0 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.2 | 4.9 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.2 | 4.4 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.2 | 5.1 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.2 | 5.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.2 | 10.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.2 | 5.1 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.2 | 2.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 2.9 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 4.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.2 | 5.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 1.9 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 2.8 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 13.5 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.2 | 19.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.5 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 10.3 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 6.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 0.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 3.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 6.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 22.0 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.1 | 0.6 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 1.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 0.8 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 1.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 3.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 8.7 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 3.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.3 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 3.9 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 1.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 1.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.8 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.9 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.9 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 1.0 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.4 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.0 | 0.6 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.6 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.9 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |