Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
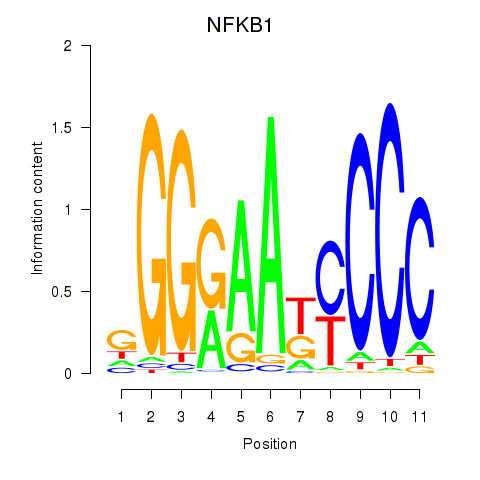
Results for NFKB1
Z-value: 1.37
Transcription factors associated with NFKB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFKB1
|
ENSG00000109320.13 | NFKB1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFKB1 | hg38_v1_chr4_+_102501885_102501957, hg38_v1_chr4_+_102501298_102501474 | 0.33 | 7.1e-07 | Click! |
Activity profile of NFKB1 motif
Sorted Z-values of NFKB1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NFKB1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 19.3 | 58.0 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 18.0 | 53.9 | GO:0072573 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) |
| 17.4 | 52.1 | GO:0035691 | macrophage migration inhibitory factor signaling pathway(GO:0035691) |
| 12.2 | 48.7 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 8.8 | 61.8 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 7.7 | 23.2 | GO:2001153 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 7.2 | 43.5 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 6.4 | 19.3 | GO:1900226 | positive regulation of oocyte maturation(GO:1900195) negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 5.2 | 15.6 | GO:1904899 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 4.6 | 13.7 | GO:2000753 | regulation of phospholipid scramblase activity(GO:1900161) positive regulation of phospholipid scramblase activity(GO:1900163) regulation of glucosylceramide catabolic process(GO:2000752) positive regulation of glucosylceramide catabolic process(GO:2000753) regulation of sphingomyelin catabolic process(GO:2000754) positive regulation of sphingomyelin catabolic process(GO:2000755) |
| 4.5 | 22.4 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 4.4 | 13.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 4.2 | 12.7 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 4.2 | 46.2 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 3.4 | 13.6 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 3.4 | 10.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 3.3 | 23.0 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 3.2 | 12.6 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 2.9 | 28.6 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 2.7 | 8.0 | GO:1990654 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 2.7 | 13.3 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 2.6 | 23.7 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 2.6 | 13.1 | GO:2000630 | positive regulation of miRNA metabolic process(GO:2000630) |
| 2.6 | 10.4 | GO:0033590 | response to cobalamin(GO:0033590) |
| 2.5 | 7.4 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 2.5 | 7.4 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 2.2 | 8.9 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 2.1 | 17.0 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 2.1 | 6.3 | GO:0009386 | translational attenuation(GO:0009386) |
| 1.9 | 5.8 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 1.9 | 13.0 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 1.8 | 5.3 | GO:2000176 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 1.7 | 20.0 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 1.7 | 29.9 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 1.4 | 7.2 | GO:0002730 | regulation of dendritic cell cytokine production(GO:0002730) |
| 1.4 | 15.5 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 1.4 | 4.1 | GO:0002876 | positive regulation of chronic inflammatory response to antigenic stimulus(GO:0002876) |
| 1.3 | 12.0 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 1.3 | 14.3 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 1.3 | 7.8 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 1.3 | 3.9 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 1.2 | 9.7 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 1.2 | 4.7 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 1.2 | 8.1 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 1.1 | 3.4 | GO:0031938 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) regulation of chromatin silencing at telomere(GO:0031938) |
| 1.1 | 3.4 | GO:1904616 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 1.1 | 10.1 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 1.1 | 4.5 | GO:0060023 | soft palate development(GO:0060023) |
| 1.1 | 14.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 1.1 | 7.5 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 1.1 | 6.3 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 1.0 | 2.9 | GO:1990451 | cellular stress response to acidic pH(GO:1990451) |
| 1.0 | 15.5 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 1.0 | 14.5 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.9 | 3.7 | GO:0052031 | tolerance induction to self antigen(GO:0002513) ossification involved in bone remodeling(GO:0043932) induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) positive regulation of mononuclear cell migration(GO:0071677) modulation by virus of host immune response(GO:0075528) negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.9 | 9.9 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.9 | 2.7 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.9 | 10.4 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.8 | 3.3 | GO:1903755 | regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 0.8 | 2.5 | GO:2000697 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 0.8 | 3.3 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.8 | 12.9 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.8 | 3.0 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.8 | 4.5 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.8 | 2.3 | GO:0071206 | establishment of protein localization to juxtaparanode region of axon(GO:0071206) |
| 0.7 | 11.1 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.7 | 2.1 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.7 | 4.8 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.7 | 5.5 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.7 | 2.7 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.7 | 2.0 | GO:0051040 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) regulation of calcium-independent cell-cell adhesion(GO:0051040) corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.6 | 3.0 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.6 | 3.6 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.6 | 14.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.5 | 27.2 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.5 | 2.7 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.5 | 4.0 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.5 | 7.9 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.5 | 1.9 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.5 | 4.7 | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) |
| 0.5 | 4.2 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.5 | 3.2 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.4 | 59.2 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.4 | 6.6 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.4 | 7.3 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.4 | 2.6 | GO:0048840 | otolith development(GO:0048840) |
| 0.4 | 3.3 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.4 | 7.9 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.4 | 24.3 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.4 | 2.3 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.4 | 4.1 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.4 | 7.7 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.4 | 2.6 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.4 | 1.5 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.4 | 0.7 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.3 | 5.0 | GO:0048535 | lymph node development(GO:0048535) |
| 0.3 | 5.3 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.3 | 1.6 | GO:0030070 | insulin processing(GO:0030070) |
| 0.3 | 1.6 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.3 | 1.9 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.3 | 1.6 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.3 | 2.7 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.3 | 0.8 | GO:0099403 | negative regulation of maintenance of sister chromatid cohesion(GO:0034092) negative regulation of maintenance of mitotic sister chromatid cohesion(GO:0034183) protein auto-ADP-ribosylation(GO:0070213) maintenance of mitotic sister chromatid cohesion, telomeric(GO:0099403) mitotic sister chromatid cohesion, telomeric(GO:0099404) regulation of telomeric DNA binding(GO:1904742) regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904907) negative regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904908) |
| 0.3 | 5.6 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.3 | 4.2 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.3 | 3.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.3 | 4.1 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.3 | 2.9 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.3 | 1.8 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.3 | 1.8 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.3 | 2.1 | GO:0043102 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.3 | 1.8 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.2 | 2.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.2 | 10.8 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.2 | 3.3 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.2 | 14.9 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.2 | 6.6 | GO:0098743 | cell aggregation(GO:0098743) |
| 0.2 | 13.9 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.2 | 1.0 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) |
| 0.2 | 5.4 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.2 | 11.8 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.2 | 1.8 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.2 | 5.7 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.2 | 5.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.2 | 10.4 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.2 | 2.3 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.2 | 4.2 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.2 | 4.2 | GO:0006833 | water transport(GO:0006833) |
| 0.2 | 83.7 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.2 | 3.8 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.2 | 12.7 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.2 | 7.9 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 17.4 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.2 | 1.0 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.2 | 4.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 0.9 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.2 | 3.6 | GO:0032754 | positive regulation of interleukin-5 production(GO:0032754) |
| 0.1 | 2.3 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.1 | 5.3 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.1 | 2.2 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 0.8 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.1 | 3.1 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.1 | 1.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 1.6 | GO:0001977 | renal system process involved in regulation of blood volume(GO:0001977) |
| 0.1 | 15.7 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 1.7 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 1.6 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 1.2 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.1 | 2.1 | GO:0050718 | positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.1 | 1.0 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 3.0 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 1.0 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 1.1 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.1 | 3.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 4.6 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.1 | 8.4 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 7.5 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.1 | 4.7 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 16.3 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 2.8 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.1 | 5.4 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.1 | 0.5 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.1 | 10.2 | GO:0090501 | RNA phosphodiester bond hydrolysis(GO:0090501) |
| 0.1 | 0.2 | GO:0051572 | negative regulation of histone H3-K36 methylation(GO:0000415) negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.1 | 11.8 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 5.5 | GO:2000257 | regulation of protein activation cascade(GO:2000257) |
| 0.0 | 1.4 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 3.0 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 4.3 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.0 | 2.0 | GO:0002456 | T cell mediated immunity(GO:0002456) |
| 0.0 | 2.2 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 5.2 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 1.1 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 0.5 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 1.7 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 3.0 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 1.2 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 1.0 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 4.3 | GO:0007162 | negative regulation of cell adhesion(GO:0007162) |
| 0.0 | 4.5 | GO:0006310 | DNA recombination(GO:0006310) |
| 0.0 | 1.5 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.5 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.7 | GO:0090003 | regulation of establishment of protein localization to plasma membrane(GO:0090003) |
| 0.0 | 0.9 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 2.5 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 1.2 | GO:0070268 | cornification(GO:0070268) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 105.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 4.9 | 63.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 3.2 | 16.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 2.6 | 28.6 | GO:0042611 | MHC protein complex(GO:0042611) |
| 2.5 | 12.7 | GO:0042825 | TAP complex(GO:0042825) |
| 2.1 | 19.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 1.8 | 12.8 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 1.7 | 22.4 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 1.5 | 4.5 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 1.5 | 10.4 | GO:0043196 | varicosity(GO:0043196) |
| 1.2 | 58.0 | GO:0097542 | ciliary tip(GO:0097542) |
| 1.1 | 3.4 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 1.1 | 3.3 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 1.0 | 4.1 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.9 | 5.6 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.9 | 5.5 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.9 | 7.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.9 | 5.3 | GO:0060091 | kinocilium(GO:0060091) |
| 0.9 | 6.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.8 | 3.3 | GO:1990356 | sumoylated E2 ligase complex(GO:1990356) |
| 0.8 | 4.0 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.7 | 10.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.7 | 2.9 | GO:1990393 | 3M complex(GO:1990393) |
| 0.7 | 123.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.7 | 49.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.7 | 2.7 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.6 | 3.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.5 | 47.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.5 | 5.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.5 | 5.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.5 | 2.7 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.4 | 17.1 | GO:0002102 | podosome(GO:0002102) |
| 0.4 | 28.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.4 | 3.0 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.4 | 14.2 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.4 | 1.9 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.4 | 7.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.4 | 2.2 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.4 | 4.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.3 | 2.8 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.3 | 7.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.3 | 37.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.3 | 27.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.3 | 1.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.3 | 1.6 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.3 | 3.0 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.3 | 2.7 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.2 | 2.8 | GO:0044613 | nuclear pore central transport channel(GO:0044613) nuclear pore nuclear basket(GO:0044615) |
| 0.2 | 2.3 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.2 | 1.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 15.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 2.9 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 26.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.2 | 2.3 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.2 | 1.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 56.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.2 | 2.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 4.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 1.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 19.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 7.3 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 4.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 5.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 3.3 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 2.9 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.1 | 9.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 14.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 7.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 16.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 6.7 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 1.0 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 8.4 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.1 | 10.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 1.0 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 6.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 7.5 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.1 | 1.4 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 16.4 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 8.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 1.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.8 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 2.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 34.7 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.1 | 8.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 1.2 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 12.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 4.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 5.8 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 1.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.6 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 6.5 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 2.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.0 | 111.9 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 7.4 | 52.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 6.6 | 39.4 | GO:0046979 | TAP2 binding(GO:0046979) |
| 6.4 | 19.3 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 4.5 | 53.6 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 3.9 | 46.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 3.7 | 51.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 2.9 | 23.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 2.9 | 14.3 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 2.5 | 9.9 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 2.3 | 13.7 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 2.0 | 8.1 | GO:0005151 | interleukin-1, Type II receptor binding(GO:0005151) |
| 1.9 | 5.6 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 1.8 | 7.4 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 1.6 | 4.7 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 1.4 | 15.6 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 1.4 | 15.5 | GO:0004568 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 1.4 | 9.7 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 1.4 | 28.7 | GO:0043495 | protein anchor(GO:0043495) |
| 1.3 | 10.4 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 1.2 | 13.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 1.2 | 4.7 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 1.2 | 3.5 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 1.1 | 4.4 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 1.1 | 7.5 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 1.0 | 19.1 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 1.0 | 3.0 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 1.0 | 2.9 | GO:0035375 | zymogen binding(GO:0035375) |
| 1.0 | 2.9 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.9 | 7.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.9 | 45.3 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.9 | 2.6 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.8 | 3.4 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.8 | 3.3 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.8 | 3.3 | GO:0050473 | arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.8 | 2.4 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.8 | 24.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.8 | 3.8 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.7 | 9.0 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.7 | 3.7 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.7 | 2.2 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.7 | 1.4 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.7 | 4.8 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.7 | 2.7 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.7 | 3.4 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.6 | 4.5 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.6 | 1.9 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.6 | 2.5 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.6 | 7.9 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.6 | 7.8 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.6 | 3.3 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.5 | 1.6 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.5 | 1.6 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.5 | 24.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.5 | 6.6 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.5 | 4.1 | GO:0046935 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.5 | 12.8 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.4 | 13.9 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.4 | 3.0 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.4 | 5.7 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.4 | 12.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.3 | 3.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.3 | 2.8 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.3 | 11.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.3 | 4.2 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.3 | 10.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.3 | 4.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.3 | 2.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.3 | 5.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.3 | 8.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.3 | 4.0 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.3 | 1.4 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.3 | 8.9 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.3 | 1.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.3 | 12.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.3 | 5.8 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.3 | 5.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.3 | 4.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.3 | 100.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.3 | 1.0 | GO:0019976 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.2 | 14.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.2 | 3.0 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.2 | 4.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.2 | 1.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 16.3 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.2 | 3.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 2.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 6.2 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.2 | 1.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 7.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 7.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.2 | 3.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 3.0 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 3.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 14.3 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.2 | 1.7 | GO:0015386 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 7.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 1.9 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 3.0 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.2 | 2.0 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 1.6 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.2 | 8.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 4.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 2.6 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.2 | 1.5 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.2 | 4.5 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 6.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 3.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 10.1 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 2.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 14.1 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 3.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 5.4 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 2.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 32.3 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.1 | 2.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 8.9 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 15.3 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 5.3 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 5.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 2.9 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 3.9 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.7 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 4.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 89.7 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.1 | 1.6 | GO:0030552 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) cAMP binding(GO:0030552) |
| 0.1 | 2.1 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 1.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 2.3 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.7 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 2.4 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 4.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 2.3 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 1.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 5.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 1.0 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 5.7 | GO:0019901 | protein kinase binding(GO:0019901) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 180.0 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 1.7 | 37.4 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 1.3 | 65.1 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 1.1 | 70.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 1.0 | 25.9 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.9 | 38.9 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.8 | 7.5 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.7 | 16.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.6 | 1.9 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.5 | 45.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.5 | 2.8 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.4 | 9.0 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.4 | 20.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.4 | 16.1 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.3 | 22.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.3 | 10.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.3 | 10.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 4.2 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.2 | 5.1 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.2 | 10.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.2 | 2.7 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.2 | 7.2 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.2 | 7.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 3.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 4.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 2.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 2.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 7.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 3.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 4.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 5.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 13.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 2.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 26.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 5.3 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 1.8 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 2.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 3.4 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 3.2 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 10.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 6.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.1 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 21.3 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 3.5 | 74.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 2.7 | 133.8 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 2.6 | 28.6 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 2.6 | 53.6 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 1.4 | 16.7 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 1.3 | 29.2 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 1.3 | 5.3 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 1.0 | 42.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.9 | 20.8 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.7 | 43.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.5 | 7.9 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.4 | 3.6 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.4 | 17.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.4 | 16.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.4 | 31.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.4 | 11.8 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.4 | 23.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.4 | 18.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.3 | 6.9 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.3 | 13.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.3 | 8.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.3 | 37.6 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.3 | 4.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 9.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.3 | 20.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.3 | 7.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.3 | 5.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.3 | 5.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.2 | 1.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 3.0 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 10.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 3.7 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 4.1 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.2 | 1.5 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 14.2 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.2 | 6.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 3.8 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 16.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 3.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 19.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 5.7 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.1 | 10.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 4.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 7.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 8.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 9.5 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 4.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 1.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 7.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 4.1 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 1.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 0.7 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 2.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.6 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 4.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.4 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 1.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 1.0 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 2.8 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 2.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 2.4 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 1.1 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 1.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |