Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
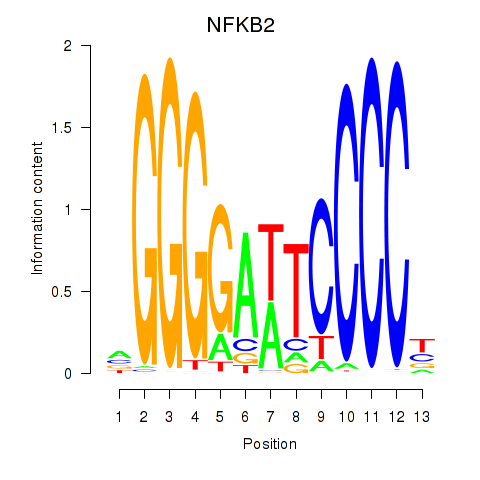
Results for NFKB2
Z-value: 1.31
Transcription factors associated with NFKB2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFKB2
|
ENSG00000077150.20 | NFKB2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFKB2 | hg38_v1_chr10_+_102395693_102395777 | 0.49 | 1.6e-14 | Click! |
Activity profile of NFKB2 motif
Sorted Z-values of NFKB2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NFKB2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.2 | 27.7 | GO:2000752 | regulation of phospholipid scramblase activity(GO:1900161) positive regulation of phospholipid scramblase activity(GO:1900163) regulation of glucosylceramide catabolic process(GO:2000752) positive regulation of glucosylceramide catabolic process(GO:2000753) regulation of sphingomyelin catabolic process(GO:2000754) positive regulation of sphingomyelin catabolic process(GO:2000755) |
| 8.5 | 25.5 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 6.6 | 26.2 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 5.6 | 16.7 | GO:0097698 | telomere maintenance via base-excision repair(GO:0097698) |
| 4.3 | 47.5 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 4.1 | 12.4 | GO:0046080 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 3.9 | 11.8 | GO:2000417 | negative regulation of eosinophil activation(GO:1902567) negative regulation of membrane invagination(GO:1905154) negative regulation of eosinophil migration(GO:2000417) |
| 3.9 | 3.9 | GO:0002442 | serotonin production involved in inflammatory response(GO:0002351) serotonin secretion involved in inflammatory response(GO:0002442) serotonin secretion by platelet(GO:0002554) |
| 3.8 | 48.8 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 3.6 | 10.9 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 3.4 | 23.5 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 3.3 | 19.8 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 2.4 | 14.6 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 2.4 | 9.6 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 2.3 | 29.7 | GO:0021769 | orbitofrontal cortex development(GO:0021769) |
| 2.3 | 18.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 2.2 | 28.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 2.1 | 10.7 | GO:0044209 | AMP salvage(GO:0044209) |
| 2.1 | 10.4 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 2.0 | 14.3 | GO:1904431 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) positive regulation of t-circle formation(GO:1904431) |
| 1.9 | 21.2 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 1.9 | 20.7 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 1.9 | 9.4 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 1.8 | 29.5 | GO:0060056 | mammary gland involution(GO:0060056) |
| 1.8 | 5.5 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 1.6 | 11.4 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 1.6 | 15.8 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 1.6 | 6.2 | GO:0075528 | ossification involved in bone remodeling(GO:0043932) induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 1.5 | 6.0 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 1.5 | 4.5 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 1.5 | 9.0 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 1.5 | 5.8 | GO:0048627 | myoblast development(GO:0048627) |
| 1.4 | 4.3 | GO:0002876 | positive regulation of chronic inflammatory response to antigenic stimulus(GO:0002876) |
| 1.4 | 9.9 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 1.4 | 7.0 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 1.4 | 9.7 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 1.3 | 5.4 | GO:0044571 | [2Fe-2S] cluster assembly(GO:0044571) |
| 1.3 | 20.9 | GO:0045116 | protein neddylation(GO:0045116) |
| 1.3 | 5.2 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 1.2 | 5.0 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 1.2 | 9.9 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 1.2 | 12.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 1.2 | 5.9 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 1.1 | 11.5 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 1.1 | 2.2 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 1.1 | 9.7 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 1.0 | 28.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 1.0 | 7.2 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 1.0 | 17.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 1.0 | 7.8 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 1.0 | 5.8 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.9 | 3.8 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.9 | 8.9 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.9 | 15.1 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.9 | 18.0 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.8 | 3.4 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.8 | 5.8 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.8 | 11.6 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.8 | 13.1 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.8 | 21.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.8 | 4.0 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.8 | 2.3 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.7 | 72.7 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.7 | 2.1 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.7 | 3.5 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.7 | 6.0 | GO:0046070 | dGTP metabolic process(GO:0046070) |
| 0.7 | 15.2 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.6 | 11.5 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.6 | 7.0 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.6 | 2.5 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.6 | 3.1 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.6 | 3.1 | GO:0015891 | response to mycotoxin(GO:0010046) iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.6 | 9.0 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.6 | 2.4 | GO:1902378 | vestibulocochlear nerve structural organization(GO:0021649) neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) endothelial tip cell fate specification(GO:0097102) positive regulation of retinal ganglion cell axon guidance(GO:1902336) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.6 | 5.3 | GO:0007068 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.6 | 4.0 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.6 | 2.3 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.6 | 2.3 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.6 | 3.9 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.5 | 3.1 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.5 | 1.6 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 0.5 | 5.7 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.5 | 10.9 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.5 | 6.7 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.5 | 9.4 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.5 | 2.9 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.5 | 3.8 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.5 | 6.1 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.5 | 24.8 | GO:0043486 | histone exchange(GO:0043486) |
| 0.5 | 1.4 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.5 | 18.6 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.4 | 78.4 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.4 | 13.3 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.4 | 2.0 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.4 | 17.4 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.4 | 2.4 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.4 | 33.9 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.4 | 1.5 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.4 | 6.0 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.4 | 2.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.3 | 3.4 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.3 | 3.4 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.3 | 3.3 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.3 | 5.2 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.3 | 1.3 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.3 | 1.9 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.3 | 1.6 | GO:0045359 | positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.3 | 3.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.3 | 3.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.3 | 5.2 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.3 | 10.2 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.3 | 1.6 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.3 | 9.7 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.3 | 1.6 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.3 | 4.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 1.8 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.2 | 2.0 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.2 | 5.5 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.2 | 1.1 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.2 | 7.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 15.3 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.2 | 6.2 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.2 | 9.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.2 | 0.6 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.2 | 2.3 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.2 | 6.3 | GO:0060065 | uterus development(GO:0060065) |
| 0.2 | 6.9 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.2 | 1.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.2 | 11.4 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.2 | 12.1 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.2 | 22.0 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.2 | 1.9 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.2 | 1.1 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.2 | 3.1 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.2 | 6.9 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.1 | 0.4 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.1 | 6.3 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 1.1 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.1 | 5.8 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 11.6 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.1 | 3.0 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 1.2 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.1 | 3.6 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 3.2 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.1 | 2.4 | GO:0050718 | positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.1 | 1.9 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.1 | 4.4 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 1.0 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 2.7 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.1 | 13.4 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.1 | 1.7 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.1 | 7.2 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.1 | 4.5 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.1 | 12.6 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.1 | 2.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.4 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.1 | 6.8 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 2.7 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 0.6 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 0.1 | 3.9 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 5.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 4.8 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 1.9 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 1.0 | GO:0046320 | regulation of fatty acid oxidation(GO:0046320) |
| 0.1 | 1.2 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.1 | 1.3 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 1.8 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.5 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.2 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.0 | GO:0098907 | regulation of SA node cell action potential(GO:0098907) positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 3.5 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.2 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.4 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 2.9 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.7 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 3.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 2.6 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 1.4 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.0 | 13.7 | GO:0043312 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 2.5 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 1.8 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 0.6 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.1 | GO:0038089 | positive regulation of cell migration by vascular endothelial growth factor signaling pathway(GO:0038089) positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 2.2 | GO:0042254 | ribosome biogenesis(GO:0042254) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.5 | 62.0 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 5.1 | 25.5 | GO:0031523 | Myb complex(GO:0031523) |
| 5.1 | 15.2 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 3.3 | 23.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 3.0 | 11.9 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 2.9 | 11.6 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 2.6 | 13.1 | GO:0032044 | DSIF complex(GO:0032044) |
| 2.5 | 10.0 | GO:0071942 | XPC complex(GO:0071942) |
| 2.2 | 9.0 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 2.1 | 8.6 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 2.0 | 12.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 2.0 | 48.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 1.9 | 36.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 1.8 | 14.3 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) ERCC4-ERCC1 complex(GO:0070522) |
| 1.7 | 8.3 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 1.6 | 29.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 1.6 | 18.9 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 1.4 | 15.8 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 1.3 | 4.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 1.3 | 2.6 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 1.2 | 5.0 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 1.2 | 10.8 | GO:0032010 | phagolysosome(GO:0032010) |
| 1.1 | 23.2 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 1.0 | 7.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.9 | 10.4 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.8 | 9.7 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.8 | 2.4 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.7 | 3.6 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.7 | 9.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.7 | 6.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.6 | 48.9 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.6 | 1.8 | GO:0034657 | GID complex(GO:0034657) |
| 0.6 | 17.5 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.6 | 5.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.6 | 1.7 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.6 | 5.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.6 | 3.9 | GO:0016589 | NURF complex(GO:0016589) |
| 0.6 | 8.9 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.6 | 10.5 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.5 | 9.0 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.5 | 2.4 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.5 | 13.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.4 | 4.4 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.4 | 6.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.4 | 1.6 | GO:1990879 | CST complex(GO:1990879) |
| 0.4 | 6.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.4 | 1.9 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.4 | 12.1 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.3 | 5.2 | GO:0032059 | bleb(GO:0032059) |
| 0.3 | 5.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.3 | 65.7 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.3 | 1.5 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.3 | 2.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.3 | 13.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.3 | 2.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.3 | 28.0 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.2 | 8.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 2.4 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.2 | 3.0 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.2 | 13.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.2 | 3.9 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.2 | 40.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 24.6 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.2 | 1.8 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.2 | 8.3 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.2 | 3.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 1.9 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.2 | 16.2 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.2 | 5.3 | GO:0071564 | SWI/SNF complex(GO:0016514) npBAF complex(GO:0071564) |
| 0.1 | 1.0 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 3.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 8.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 5.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 10.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 9.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 5.7 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 16.5 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 0.1 | 3.5 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 3.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 5.3 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.7 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 5.1 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.1 | 9.0 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 38.4 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 23.1 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.1 | 2.7 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 4.0 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.1 | 1.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 2.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 5.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 5.0 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 2.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.2 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 7.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 1.3 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 2.4 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 3.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 70.0 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 17.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.9 | 62.0 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 8.7 | 43.5 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 4.9 | 14.6 | GO:0047315 | kynurenine-glyoxylate transaminase activity(GO:0047315) |
| 4.6 | 27.7 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 4.3 | 26.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 4.1 | 12.4 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 3.7 | 26.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 3.6 | 18.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 3.2 | 9.7 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 3.0 | 8.9 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 2.9 | 8.6 | GO:0035730 | S-nitrosoglutathione binding(GO:0035730) dinitrosyl-iron complex binding(GO:0035731) |
| 2.7 | 10.7 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 2.6 | 36.0 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 2.5 | 15.0 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 2.2 | 48.8 | GO:0008494 | translation activator activity(GO:0008494) |
| 1.9 | 9.6 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 1.9 | 9.4 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 1.8 | 5.5 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 1.8 | 7.2 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 1.8 | 14.3 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 1.7 | 11.9 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 1.5 | 39.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 1.5 | 6.0 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 1.4 | 5.6 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 1.4 | 5.5 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 1.3 | 10.8 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 1.3 | 4.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 1.2 | 6.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 1.2 | 5.8 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 1.1 | 11.4 | GO:0050692 | DBD domain binding(GO:0050692) |
| 1.1 | 21.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 1.1 | 20.9 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 1.0 | 41.2 | GO:0019956 | chemokine binding(GO:0019956) |
| 1.0 | 8.8 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.9 | 15.8 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.9 | 4.5 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.9 | 28.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.9 | 5.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.8 | 11.8 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.8 | 10.9 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.8 | 14.7 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.8 | 35.5 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.8 | 7.8 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.8 | 19.6 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.7 | 4.4 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.7 | 8.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.7 | 28.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.7 | 2.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.7 | 6.0 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.6 | 7.0 | GO:0004568 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.6 | 6.3 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.6 | 1.9 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.6 | 9.0 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.6 | 3.6 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.6 | 7.0 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.5 | 9.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.5 | 3.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.5 | 4.4 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.5 | 20.7 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.5 | 98.5 | GO:0003823 | antigen binding(GO:0003823) |
| 0.5 | 17.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.4 | 4.5 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.4 | 27.5 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.4 | 1.6 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.4 | 6.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.4 | 5.7 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.4 | 18.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.4 | 71.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.4 | 1.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.4 | 2.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.4 | 5.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.4 | 3.9 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.3 | 10.8 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.3 | 1.0 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.3 | 3.0 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.3 | 10.4 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.3 | 3.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.3 | 6.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.3 | 1.5 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.3 | 11.6 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.3 | 2.4 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.3 | 7.5 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.3 | 1.9 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.3 | 3.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.3 | 9.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.3 | 14.5 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.2 | 1.5 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.2 | 10.9 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.2 | 3.9 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 1.7 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.2 | 65.5 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.2 | 5.4 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.2 | 1.6 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.2 | 4.4 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.2 | 1.9 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.2 | 1.9 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 26.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 19.6 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.2 | 10.0 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.2 | 3.3 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 1.0 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 4.0 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.4 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.1 | 3.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 1.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 4.7 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 3.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 4.0 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 2.4 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.1 | 35.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 0.5 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.8 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 5.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 3.4 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 3.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 1.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 2.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 3.5 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 2.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 2.0 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 1.8 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 3.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 19.5 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 3.9 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 1.9 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 3.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.2 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 2.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.4 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.0 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 1.3 | 65.4 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 1.0 | 18.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.8 | 25.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.8 | 36.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.7 | 69.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.6 | 37.0 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.5 | 7.9 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.5 | 19.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.4 | 18.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.4 | 26.2 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.4 | 8.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.4 | 4.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.3 | 5.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.3 | 16.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.3 | 13.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.3 | 10.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.3 | 16.8 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.3 | 9.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.3 | 6.4 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.2 | 10.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 14.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 5.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 3.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 10.4 | PID FOXO PATHWAY | FoxO family signaling |
| 0.2 | 2.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 13.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 3.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 1.4 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 8.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 4.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 4.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 2.5 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 1.1 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 1.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 2.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 2.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 5.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 3.1 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 7.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 3.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 2.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 8.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 26.0 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 1.7 | 46.4 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 1.6 | 58.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 1.3 | 21.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 1.2 | 38.6 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 1.1 | 45.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 1.0 | 15.0 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.9 | 80.2 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.9 | 60.3 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.9 | 26.1 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.8 | 18.0 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.8 | 16.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.7 | 24.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.7 | 25.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.6 | 9.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.6 | 17.4 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.6 | 11.9 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.5 | 17.4 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.5 | 11.1 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.5 | 9.9 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.5 | 9.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.4 | 5.8 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.4 | 9.9 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.4 | 3.9 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.4 | 18.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.4 | 13.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.4 | 13.3 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.4 | 6.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.4 | 12.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.4 | 6.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 5.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.3 | 9.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.3 | 5.0 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.3 | 6.1 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.3 | 5.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 8.4 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.3 | 2.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.3 | 7.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.3 | 10.8 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.3 | 3.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.3 | 4.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.3 | 8.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 4.5 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.2 | 7.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.2 | 1.6 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.2 | 3.5 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.2 | 2.7 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 3.5 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.2 | 2.4 | REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | Genes involved in Nucleotide-binding domain, leucine rich repeat containing receptor (NLR) signaling pathways |
| 0.2 | 3.9 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 4.5 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.2 | 5.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.2 | 8.2 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.2 | 15.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 5.5 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.1 | 3.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 3.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 5.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 6.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 4.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 7.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.5 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.1 | 13.9 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.1 | 1.0 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 2.0 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.1 | 1.1 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 2.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 4.2 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.6 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.9 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.1 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |