Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
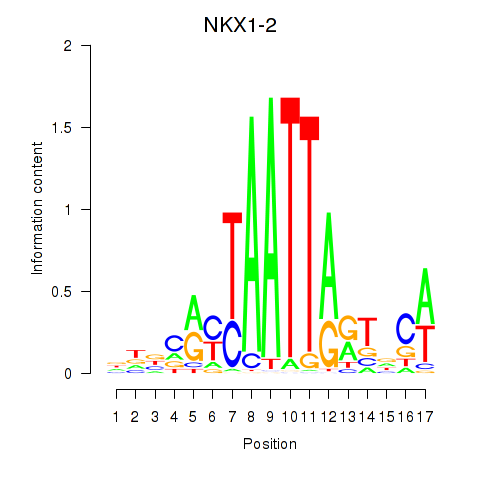
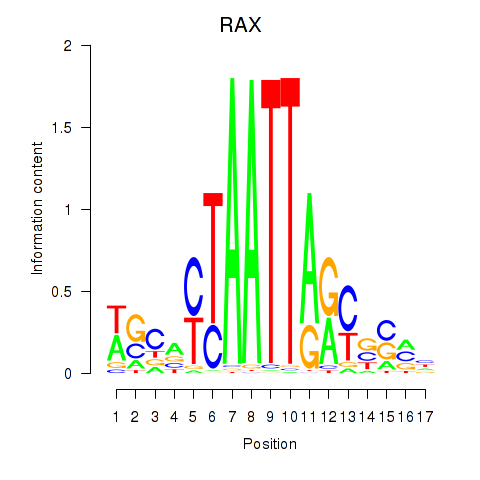
Results for NKX1-2_RAX
Z-value: 0.63
Transcription factors associated with NKX1-2_RAX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX1-2
|
ENSG00000229544.9 | NKX1-2 |
|
RAX
|
ENSG00000134438.10 | RAX |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RAX | hg38_v1_chr18_-_59273447_59273459, hg38_v1_chr18_-_59273379_59273429 | -0.55 | 7.8e-19 | Click! |
Activity profile of NKX1-2_RAX motif
Sorted Z-values of NKX1-2_RAX motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX1-2_RAX
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 13.2 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) mitochondrial membrane fission(GO:0090149) |
| 4.3 | 30.4 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 2.4 | 11.8 | GO:0045337 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 1.6 | 4.7 | GO:0001300 | chronological cell aging(GO:0001300) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.9 | 2.8 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.9 | 2.7 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.8 | 16.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.8 | 6.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.7 | 28.8 | GO:0071173 | mitotic spindle assembly checkpoint(GO:0007094) spindle assembly checkpoint(GO:0071173) |
| 0.7 | 2.1 | GO:0031455 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.7 | 32.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.7 | 2.0 | GO:0071284 | copper ion export(GO:0060003) cellular response to lead ion(GO:0071284) regulation of electron carrier activity(GO:1904732) |
| 0.7 | 2.0 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.6 | 4.5 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.4 | 3.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.4 | 5.7 | GO:0042407 | cristae formation(GO:0042407) |
| 0.4 | 3.6 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.4 | 1.9 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.4 | 13.0 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.3 | 2.7 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.3 | 7.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.3 | 4.5 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.3 | 6.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.3 | 1.2 | GO:0030047 | actin modification(GO:0030047) |
| 0.3 | 3.7 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.3 | 5.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 1.8 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.2 | 18.5 | GO:0051439 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) |
| 0.2 | 4.0 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.2 | 1.6 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.2 | 2.4 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 2.8 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.1 | 0.9 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 2.0 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 1.6 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 0.5 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.1 | 3.0 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.1 | 2.2 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 2.8 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.1 | 3.0 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 2.3 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.1 | 2.4 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.1 | 5.8 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.1 | 2.2 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.1 | 5.6 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 2.3 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.1 | 0.5 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 1.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 2.9 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.4 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.4 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 1.3 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 1.1 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.5 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 1.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 3.3 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 1.6 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 1.3 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 1.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.4 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 1.9 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 3.0 | GO:1901796 | regulation of signal transduction by p53 class mediator(GO:1901796) |
| 0.0 | 0.8 | GO:0002456 | T cell mediated immunity(GO:0002456) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:2000782 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.6 | 28.8 | GO:1990298 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 4.3 | 30.4 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 1.3 | 4.0 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.9 | 5.7 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.9 | 3.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.8 | 3.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.7 | 3.0 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.7 | 2.2 | GO:0071756 | IgM immunoglobulin complex(GO:0071753) IgM immunoglobulin complex, circulating(GO:0071754) pentameric IgM immunoglobulin complex(GO:0071756) |
| 0.6 | 18.5 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.4 | 3.6 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.4 | 4.5 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.4 | 1.6 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.4 | 2.8 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.4 | 1.9 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.4 | 1.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.3 | 5.1 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.3 | 1.2 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.3 | 5.8 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.2 | 4.5 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 1.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 3.0 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 13.2 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 5.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 1.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 9.0 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 1.6 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 1.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 7.6 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 2.0 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 3.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 4.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 25.3 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 2.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 3.8 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.4 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 2.7 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 2.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 3.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 5.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 2.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 4.1 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 16.3 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 2.4 | 11.8 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 1.3 | 6.7 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 1.3 | 6.3 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.9 | 2.7 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.9 | 3.5 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.8 | 2.4 | GO:0000252 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) sterol-4-alpha-carboxylate 3-dehydrogenase (decarboxylating) activity(GO:0047012) |
| 0.8 | 34.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.8 | 30.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.7 | 13.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.7 | 2.0 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.7 | 2.0 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) phosphoglycerate kinase activity(GO:0004618) copper-transporting ATPase activity(GO:0043682) |
| 0.6 | 3.0 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.6 | 1.8 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.4 | 2.8 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.3 | 4.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.3 | 1.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.3 | 1.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.3 | 3.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.2 | 2.8 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 5.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 33.0 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.2 | 5.8 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.2 | 1.9 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.2 | 2.0 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.2 | 3.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.2 | 3.7 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 3.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 0.9 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.2 | 5.1 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 2.0 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 1.4 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 2.4 | GO:0051430 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 2.2 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.4 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 2.9 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 1.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 3.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 4.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 2.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 2.1 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 3.6 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 1.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.5 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.4 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.1 | 2.0 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 0.6 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 4.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 7.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 4.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 5.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 13.0 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 13.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 4.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 34.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.2 | 33.7 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 4.0 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 3.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 1.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 4.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 4.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 2.0 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 2.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 33.0 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 1.0 | 34.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.4 | 14.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.3 | 18.5 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.2 | 6.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.2 | 4.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 3.5 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 13.2 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.1 | 2.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 4.7 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 0.9 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 2.4 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 5.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 5.1 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 2.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 5.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 1.9 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 3.0 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 2.3 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 2.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 4.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 2.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 5.6 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 2.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |