Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for NKX6-3
Z-value: 0.30
Transcription factors associated with NKX6-3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX6-3
|
ENSG00000165066.13 | NKX6-3 |
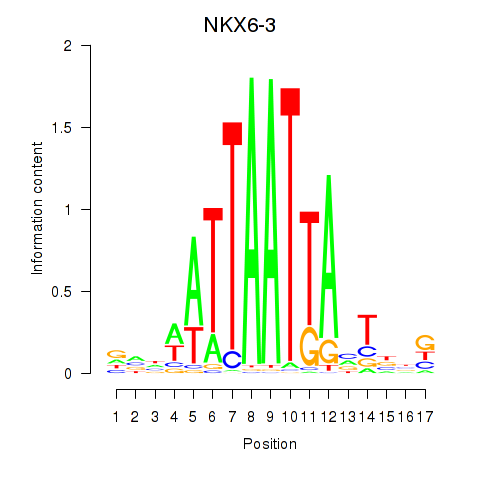
Activity profile of NKX6-3 motif
Sorted Z-values of NKX6-3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX6-3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_122370523 | 5.19 |
ENST00000643810.1
ENST00000540753.6 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr9_+_122371036 | 4.33 |
ENST00000619306.5
ENST00000426608.6 ENST00000223423.8 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr9_+_122371014 | 2.85 |
ENST00000362012.7
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr19_+_49513353 | 2.64 |
ENST00000596975.5
|
FCGRT
|
Fc fragment of IgG receptor and transporter |
| chr3_-_142029108 | 2.61 |
ENST00000497579.5
|
TFDP2
|
transcription factor Dp-2 |
| chr7_-_25228485 | 2.24 |
ENST00000222674.2
|
NPVF
|
neuropeptide VF precursor |
| chr11_-_13495984 | 2.22 |
ENST00000282091.6
|
PTH
|
parathyroid hormone |
| chr20_-_35147285 | 2.00 |
ENST00000374491.3
ENST00000374492.8 |
EDEM2
|
ER degradation enhancing alpha-mannosidase like protein 2 |
| chr11_-_13496018 | 1.96 |
ENST00000529816.1
|
PTH
|
parathyroid hormone |
| chr20_+_45416551 | 1.73 |
ENST00000639292.1
|
PIGT
|
phosphatidylinositol glycan anchor biosynthesis class T |
| chr5_-_143400716 | 1.55 |
ENST00000424646.6
ENST00000652686.1 |
NR3C1
|
nuclear receptor subfamily 3 group C member 1 |
| chr14_-_50561119 | 1.50 |
ENST00000555216.5
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr3_+_138621225 | 1.34 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr13_+_53028806 | 1.01 |
ENST00000219022.3
|
OLFM4
|
olfactomedin 4 |
| chr17_-_10469558 | 0.94 |
ENST00000255381.2
|
MYH4
|
myosin heavy chain 4 |
| chr3_-_142000353 | 0.82 |
ENST00000499676.5
|
TFDP2
|
transcription factor Dp-2 |
| chr5_-_58999885 | 0.82 |
ENST00000317118.12
|
PDE4D
|
phosphodiesterase 4D |
| chr3_+_138621207 | 0.82 |
ENST00000464668.5
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr1_-_113871665 | 0.77 |
ENST00000528414.5
ENST00000460620.5 ENST00000359785.10 ENST00000420377.6 ENST00000525799.1 ENST00000538253.5 |
PTPN22
|
protein tyrosine phosphatase non-receptor type 22 |
| chr5_-_140346596 | 0.74 |
ENST00000230990.7
|
HBEGF
|
heparin binding EGF like growth factor |
| chr15_+_48191648 | 0.70 |
ENST00000646012.1
ENST00000561127.5 ENST00000647546.1 ENST00000559641.5 ENST00000417307.3 |
SLC12A1
CTXN2
|
solute carrier family 12 member 1 cortexin 2 |
| chr6_+_113857333 | 0.54 |
ENST00000612661.2
|
MARCKS
|
myristoylated alanine rich protein kinase C substrate |
| chr19_+_4402615 | 0.42 |
ENST00000301280.10
|
CHAF1A
|
chromatin assembly factor 1 subunit A |
| chr3_+_136930469 | 0.42 |
ENST00000469404.1
ENST00000467911.1 |
NCK1
|
NCK adaptor protein 1 |
| chr15_+_92900189 | 0.40 |
ENST00000626874.2
ENST00000627622.1 ENST00000629346.2 ENST00000628375.2 ENST00000420239.7 ENST00000394196.9 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr12_-_52367478 | 0.35 |
ENST00000257901.7
|
KRT85
|
keratin 85 |
| chrX_+_108045050 | 0.32 |
ENST00000458383.1
ENST00000217957.10 |
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr4_-_122621011 | 0.32 |
ENST00000611104.2
ENST00000648588.1 |
IL21
|
interleukin 21 |
| chr17_-_66229380 | 0.29 |
ENST00000205948.11
|
APOH
|
apolipoprotein H |
| chrX_+_108044967 | 0.26 |
ENST00000415430.7
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr3_-_165078480 | 0.21 |
ENST00000264382.8
|
SI
|
sucrase-isomaltase |
| chr20_-_7940444 | 0.18 |
ENST00000378789.4
|
HAO1
|
hydroxyacid oxidase 1 |
| chr16_+_86566821 | 0.17 |
ENST00000649859.1
|
FOXC2
|
forkhead box C2 |
| chr7_+_138460238 | 0.17 |
ENST00000343526.9
|
TRIM24
|
tripartite motif containing 24 |
| chr16_-_67483541 | 0.12 |
ENST00000290953.3
|
AGRP
|
agouti related neuropeptide |
| chrX_+_43656289 | 0.06 |
ENST00000338702.4
|
MAOA
|
monoamine oxidase A |
| chr8_+_76681208 | 0.03 |
ENST00000651372.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr2_+_233917371 | 0.01 |
ENST00000324695.9
ENST00000433712.6 |
TRPM8
|
transient receptor potential cation channel subfamily M member 8 |
| chrX_+_47836877 | 0.01 |
ENST00000338637.13
ENST00000376954.6 |
ZNF81
|
zinc finger protein 81 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.2 | GO:1900158 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.9 | 2.6 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.7 | 2.0 | GO:0036510 | trimming of terminal mannose on C branch(GO:0036510) |
| 0.6 | 12.4 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.2 | 1.7 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.2 | 2.2 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.2 | 1.5 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.2 | 0.8 | GO:1902523 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.1 | 0.4 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.1 | 0.7 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.1 | 0.8 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 1.0 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.1 | 3.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.2 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.1 | 1.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.4 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.3 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.6 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.2 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.3 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.0 | 0.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.2 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.9 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 12.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 0.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 2.0 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.5 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.0 | 0.9 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 3.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 1.0 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.7 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 12.4 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.7 | 2.6 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.4 | 1.7 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.3 | 1.5 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.2 | 2.0 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 0.7 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.2 | 4.2 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 1.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.4 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.2 | GO:0052852 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 0.0 | 0.3 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.2 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.4 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.8 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.9 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 12.4 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 2.0 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.7 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 4.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.7 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |