Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
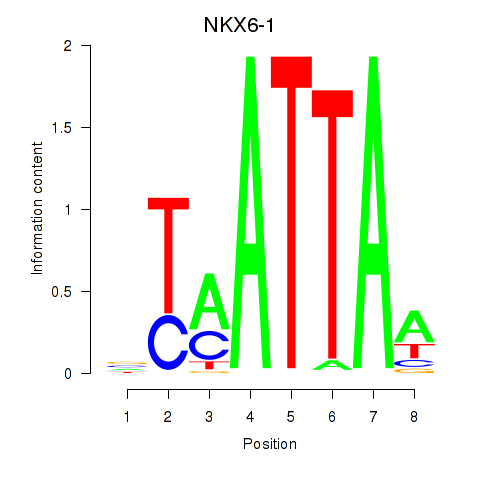
Results for NOTO_VSX2_DLX2_DLX6_NKX6-1
Z-value: 0.74
Transcription factors associated with NOTO_VSX2_DLX2_DLX6_NKX6-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
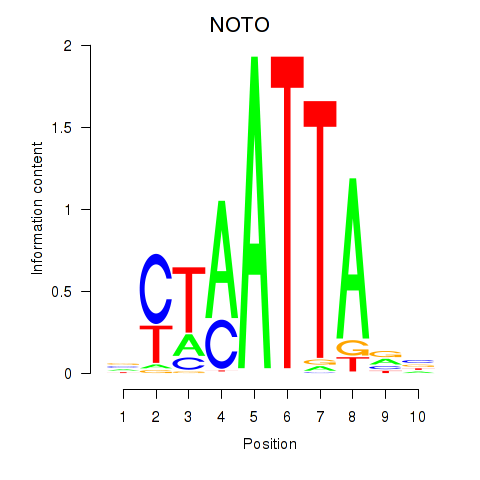
NOTO
|
ENSG00000214513.4 | NOTO |
|
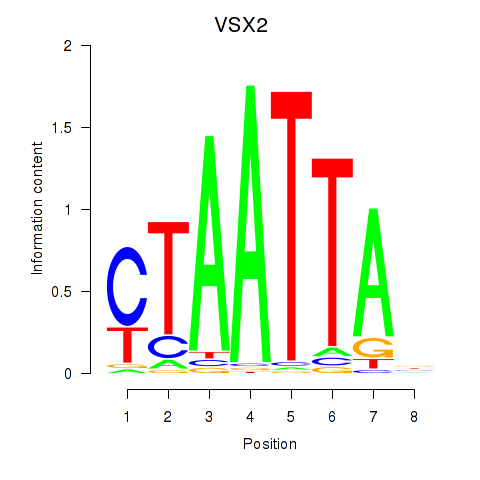
VSX2
|
ENSG00000119614.3 | VSX2 |
|
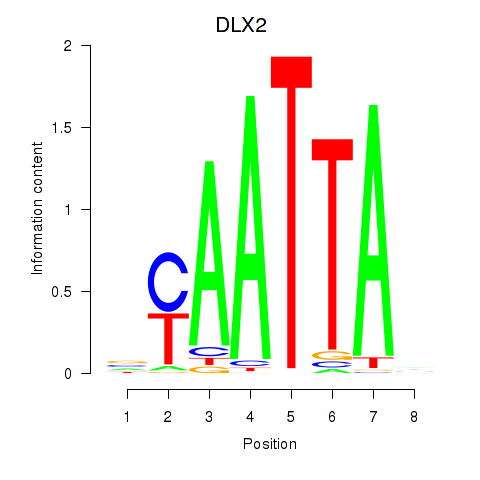
DLX2
|
ENSG00000115844.11 | DLX2 |
|
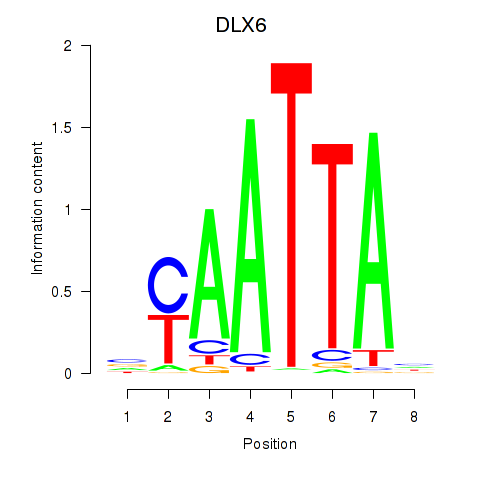
DLX6
|
ENSG00000006377.11 | DLX6 |
|
NKX6-1
|
ENSG00000163623.10 | NKX6-1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DLX6 | hg38_v1_chr7_+_97005538_97005562 | 0.47 | 1.9e-13 | Click! |
| NKX6-1 | hg38_v1_chr4_-_84499281_84499298 | 0.37 | 1.7e-08 | Click! |
| DLX2 | hg38_v1_chr2_-_172102893_172102909 | 0.19 | 4.9e-03 | Click! |
| NOTO | hg38_v1_chr2_+_73202570_73202577 | 0.10 | 1.4e-01 | Click! |
Activity profile of NOTO_VSX2_DLX2_DLX6_NKX6-1 motif
Sorted Z-values of NOTO_VSX2_DLX2_DLX6_NKX6-1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NOTO_VSX2_DLX2_DLX6_NKX6-1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.2 | 78.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 4.3 | 42.6 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 2.2 | 6.7 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 1.8 | 5.4 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 1.6 | 8.2 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 1.5 | 15.5 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 1.5 | 6.0 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 1.3 | 1.3 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 1.3 | 3.9 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 1.2 | 3.7 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 1.1 | 3.4 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 1.1 | 12.0 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 1.0 | 4.0 | GO:1900158 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.9 | 2.8 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.9 | 11.0 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.9 | 75.6 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.9 | 7.8 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.9 | 2.6 | GO:1904397 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.9 | 2.6 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.8 | 3.2 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.8 | 3.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.8 | 5.4 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.7 | 2.1 | GO:1990418 | response to insulin-like growth factor stimulus(GO:1990418) |
| 0.7 | 2.0 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.7 | 2.0 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) |
| 0.6 | 2.5 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.6 | 7.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.6 | 4.2 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.6 | 2.3 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.6 | 2.8 | GO:0070778 | L-aspartate transport(GO:0070778) L-aspartate transmembrane transport(GO:0089712) |
| 0.5 | 1.6 | GO:1904432 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.5 | 14.6 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.5 | 3.2 | GO:0050955 | thermoception(GO:0050955) |
| 0.5 | 1.6 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.5 | 7.7 | GO:0035878 | nail development(GO:0035878) |
| 0.5 | 2.0 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.5 | 3.0 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.5 | 1.5 | GO:0071934 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.5 | 2.0 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.5 | 3.4 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.5 | 10.2 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.5 | 1.4 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.5 | 3.2 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.5 | 1.4 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.4 | 1.3 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.4 | 5.0 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.4 | 1.6 | GO:0060023 | soft palate development(GO:0060023) |
| 0.4 | 0.4 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) |
| 0.4 | 1.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.4 | 1.1 | GO:1901964 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm development(GO:0048389) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) pattern specification involved in mesonephros development(GO:0061227) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in kidney development(GO:0072098) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter urothelium development(GO:0072190) ureter epithelial cell differentiation(GO:0072192) ureter morphogenesis(GO:0072197) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.3 | 1.7 | GO:0010748 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.3 | 9.3 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.3 | 2.3 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.3 | 1.3 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.3 | 3.3 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.3 | 4.8 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.3 | 0.9 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.3 | 1.2 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.3 | 1.9 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.3 | 2.4 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.3 | 1.9 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.3 | 2.0 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.2 | 3.4 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.2 | 1.0 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.2 | 13.0 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.2 | 2.8 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.2 | 1.2 | GO:0021764 | amygdala development(GO:0021764) |
| 0.2 | 3.0 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 1.7 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.2 | 7.0 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.2 | 0.8 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.2 | 0.8 | GO:0002331 | pre-B cell differentiation(GO:0002329) pre-B cell allelic exclusion(GO:0002331) |
| 0.2 | 1.2 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.2 | 1.4 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.2 | 2.3 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.2 | 0.6 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.2 | 0.2 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.2 | 1.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.2 | 1.9 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.2 | 0.3 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.2 | 1.3 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.2 | 2.8 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.2 | 0.9 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.2 | 3.7 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.1 | 0.4 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 3.6 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.1 | 2.7 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.5 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.1 | 2.2 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.1 | 0.6 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 2.4 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 1.5 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 1.1 | GO:0060125 | habituation(GO:0046959) negative regulation of growth hormone secretion(GO:0060125) |
| 0.1 | 1.4 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.2 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.1 | 2.0 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 5.5 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.1 | 3.8 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 1.0 | GO:0015868 | purine nucleotide transport(GO:0015865) purine ribonucleotide transport(GO:0015868) |
| 0.1 | 0.7 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 1.1 | GO:0015810 | aspartate transport(GO:0015810) |
| 0.1 | 2.5 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 1.3 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.1 | 1.9 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.1 | 0.4 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.1 | 0.5 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.1 | 0.4 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.5 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 2.8 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 2.2 | GO:0034776 | response to histamine(GO:0034776) |
| 0.1 | 1.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 2.7 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 2.6 | GO:0009083 | branched-chain amino acid metabolic process(GO:0009081) branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 1.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.7 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 0.3 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.9 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) myoblast fate commitment(GO:0048625) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 1.4 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 1.2 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.1 | 5.7 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.1 | 3.0 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.3 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.9 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.3 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.1 | 1.2 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.9 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 1.5 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.6 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 10.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.5 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.6 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 3.9 | GO:0007595 | lactation(GO:0007595) |
| 0.1 | 0.5 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 3.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 1.6 | GO:0007616 | long-term memory(GO:0007616) |
| 0.1 | 1.3 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.1 | 1.5 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.3 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.7 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 1.4 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.1 | 3.7 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 | 2.5 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.1 | 1.7 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 0.8 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.4 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 3.3 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 2.5 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 1.6 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 1.4 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 1.8 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 18.6 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.3 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 1.6 | GO:0009798 | axis specification(GO:0009798) |
| 0.0 | 5.1 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.0 | 43.9 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 1.7 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.2 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 3.7 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 2.5 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 2.1 | GO:0006378 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 2.1 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.6 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 2.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.5 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.1 | GO:0032908 | transforming growth factor beta1 production(GO:0032905) regulation of transforming growth factor beta1 production(GO:0032908) |
| 0.0 | 1.0 | GO:0042551 | neuron maturation(GO:0042551) |
| 0.0 | 3.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 1.6 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 0.6 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.4 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 1.6 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 0.9 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 1.9 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.6 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.8 | GO:0060964 | regulation of gene silencing by miRNA(GO:0060964) |
| 0.0 | 0.2 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.1 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 0.4 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.1 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.0 | 1.0 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.5 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.7 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 1.3 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 0.9 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.3 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 2.1 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 5.2 | GO:0019216 | regulation of lipid metabolic process(GO:0019216) |
| 0.0 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.2 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.7 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.3 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.0 | 1.0 | GO:0050729 | positive regulation of inflammatory response(GO:0050729) |
| 0.0 | 2.4 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.8 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 1.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 1.8 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
| 0.0 | 0.4 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.9 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 17.0 | GO:0071953 | elastic fiber(GO:0071953) |
| 1.0 | 4.1 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.9 | 6.0 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.8 | 45.6 | GO:0031430 | M band(GO:0031430) |
| 0.6 | 3.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.5 | 1.6 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.5 | 6.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.5 | 1.5 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.5 | 1.4 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.4 | 6.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.4 | 1.7 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.4 | 3.3 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.4 | 8.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.4 | 0.4 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.3 | 94.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.3 | 5.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.3 | 1.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.3 | 2.0 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.3 | 1.6 | GO:1990357 | terminal web(GO:1990357) |
| 0.3 | 2.7 | GO:0097433 | dense body(GO:0097433) |
| 0.3 | 1.9 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.3 | 5.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 1.4 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 1.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.2 | 2.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 4.1 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.2 | 4.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 8.6 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.2 | 1.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 9.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 3.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 1.4 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.2 | 1.0 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.1 | 3.0 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 1.6 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 2.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 1.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 14.6 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 1.7 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 12.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 21.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 2.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 2.0 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 2.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.6 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 10.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.9 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 2.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.9 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 3.6 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 0.7 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 1.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 3.3 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.1 | 0.5 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.5 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 2.8 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 1.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 2.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 1.1 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 3.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 3.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 2.9 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 2.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.0 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 2.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 63.8 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 0.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 4.0 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 1.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 2.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.1 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 1.2 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 1.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 14.7 | GO:0044432 | endoplasmic reticulum part(GO:0044432) |
| 0.0 | 1.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 1.7 | 6.7 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 1.4 | 8.2 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 1.3 | 5.4 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 1.2 | 78.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 1.0 | 4.1 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 1.0 | 3.0 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 1.0 | 3.9 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.9 | 12.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.9 | 6.0 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.8 | 6.6 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.7 | 44.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.7 | 3.9 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.6 | 2.6 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.6 | 2.5 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.6 | 8.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.6 | 2.8 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.5 | 15.4 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.5 | 1.6 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.5 | 1.6 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.5 | 12.0 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.5 | 1.5 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.5 | 2.0 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.5 | 2.8 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.5 | 1.4 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.4 | 5.4 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.4 | 6.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.4 | 1.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.4 | 2.3 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.4 | 1.1 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.3 | 3.5 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.3 | 3.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.3 | 78.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.3 | 1.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.3 | 3.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.3 | 1.0 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.3 | 7.9 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.3 | 2.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.3 | 2.8 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.2 | 1.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.2 | 2.7 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.2 | 1.5 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.2 | 0.7 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.2 | 1.4 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 5.4 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.2 | 1.9 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 3.0 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.2 | 1.6 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.2 | 1.3 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.2 | 2.0 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.2 | 6.9 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.2 | 2.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 1.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 1.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.8 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 0.4 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.8 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.4 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.1 | 1.6 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 1.0 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 3.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 3.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 3.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 1.3 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 10.9 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 5.1 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 19.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.6 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.1 | 7.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 10.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 3.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.3 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.1 | 0.4 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 2.7 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 2.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.3 | GO:0004773 | steryl-sulfatase activity(GO:0004773) |
| 0.1 | 1.0 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 21.0 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.1 | 1.3 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 6.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 2.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.6 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 13.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 1.7 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.1 | 0.7 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 1.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 2.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.5 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 0.5 | GO:0015386 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 1.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.8 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 1.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.7 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 1.4 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) sialic acid binding(GO:0033691) |
| 0.0 | 1.7 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.0 | 0.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 3.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.9 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 1.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 2.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.5 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.7 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 4.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 2.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 3.2 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 3.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.0 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 2.7 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 3.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 2.2 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 5.0 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 2.2 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0001093 | TFIIB-class transcription factor binding(GO:0001093) |
| 0.0 | 1.1 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.7 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.4 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.6 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) acetylcholine binding(GO:0042166) |
| 0.0 | 2.4 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.9 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 1.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.4 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 4.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 1.7 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 9.8 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 3.4 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.5 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.2 | GO:0001223 | transcription coactivator binding(GO:0001223) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.2 | 26.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.2 | 5.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 39.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 3.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 7.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 11.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 8.2 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 1.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 2.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 13.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 1.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 0.7 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 3.4 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 1.1 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 5.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 3.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 1.5 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 1.8 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 2.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 2.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 2.2 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 6.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 1.3 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.3 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 1.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 3.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 14.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.9 | 15.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.4 | 8.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.3 | 6.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 9.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 3.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 4.1 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 17.5 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.2 | 8.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 3.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 2.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.2 | 3.9 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.2 | 1.7 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 1.7 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.6 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 5.2 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 5.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 0.9 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 19.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 3.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.7 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 1.8 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 0.9 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 1.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 2.7 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 1.0 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 0.7 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 1.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.2 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 3.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 0.7 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 0.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 6.8 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 3.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 1.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.1 | REACTOME CD28 CO STIMULATION | Genes involved in CD28 co-stimulation |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 2.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.6 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 2.2 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.3 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.0 | 1.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 2.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.6 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 1.6 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 1.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.3 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |