Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
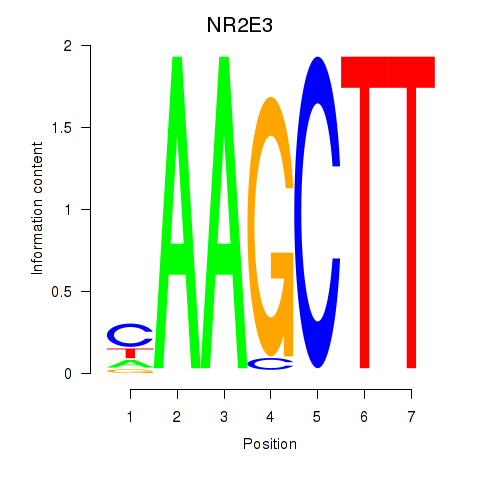
Results for NR2E3
Z-value: 1.33
Transcription factors associated with NR2E3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2E3
|
ENSG00000278570.5 | NR2E3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2E3 | hg38_v1_chr15_+_71810539_71810592 | 0.04 | 5.6e-01 | Click! |
Activity profile of NR2E3 motif
Sorted Z-values of NR2E3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2E3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 21.8 | 65.4 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 10.1 | 30.3 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 9.2 | 82.5 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 8.2 | 24.7 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 7.9 | 23.6 | GO:1904395 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 4.9 | 24.5 | GO:0035900 | response to isolation stress(GO:0035900) |
| 4.2 | 12.6 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 4.0 | 23.7 | GO:0051563 | microglia differentiation(GO:0014004) microglia development(GO:0014005) collateral sprouting in absence of injury(GO:0048669) smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) cellular response to norepinephrine stimulus(GO:0071874) |
| 3.6 | 21.6 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 3.5 | 20.9 | GO:0097338 | response to clozapine(GO:0097338) |
| 3.2 | 9.6 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 3.1 | 119.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 2.8 | 33.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 2.6 | 7.7 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 2.6 | 10.2 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 2.2 | 22.4 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 2.2 | 23.9 | GO:0097106 | postsynaptic density organization(GO:0097106) postsynaptic density assembly(GO:0097107) gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 1.9 | 9.5 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 1.9 | 5.7 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 1.8 | 7.1 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 1.6 | 16.5 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 1.6 | 19.5 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.6 | 4.9 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 1.6 | 16.0 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) |
| 1.5 | 7.3 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 1.3 | 4.0 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 1.2 | 17.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 1.2 | 3.5 | GO:1902955 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 1.1 | 4.6 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 1.1 | 4.5 | GO:0042441 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 1.1 | 4.4 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 1.1 | 28.4 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 1.1 | 5.4 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 1.0 | 13.5 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 1.0 | 5.0 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 1.0 | 4.8 | GO:1904764 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 1.0 | 7.7 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.9 | 6.4 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.9 | 5.3 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.9 | 5.1 | GO:0035602 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) |
| 0.8 | 8.9 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.8 | 10.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.8 | 7.5 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.7 | 10.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.7 | 8.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.7 | 9.3 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.7 | 12.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.7 | 5.9 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.6 | 7.1 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.6 | 6.3 | GO:1901142 | insulin metabolic process(GO:1901142) |
| 0.6 | 1.8 | GO:0060957 | endocardial cell fate commitment(GO:0060957) endocardial cushion cell fate commitment(GO:0061445) |
| 0.6 | 4.8 | GO:0030091 | protein repair(GO:0030091) |
| 0.6 | 6.4 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.6 | 5.2 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.5 | 3.8 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.5 | 1.5 | GO:0099404 | negative regulation of maintenance of sister chromatid cohesion(GO:0034092) negative regulation of maintenance of mitotic sister chromatid cohesion(GO:0034183) maintenance of mitotic sister chromatid cohesion, telomeric(GO:0099403) mitotic sister chromatid cohesion, telomeric(GO:0099404) regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904907) negative regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904908) |
| 0.5 | 12.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.5 | 1.9 | GO:1903971 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 0.5 | 2.3 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.4 | 3.0 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.4 | 0.4 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.4 | 4.3 | GO:0007549 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.4 | 1.7 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.4 | 7.9 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.4 | 3.7 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.4 | 1.2 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.4 | 9.2 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.4 | 0.8 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) |
| 0.4 | 1.5 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.4 | 4.8 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.4 | 19.0 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.3 | 1.0 | GO:0036166 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.3 | 3.0 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.3 | 3.3 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.3 | 0.9 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.3 | 3.1 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) regulation of synaptic vesicle recycling(GO:1903421) |
| 0.3 | 0.9 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.3 | 4.5 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.3 | 48.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.3 | 2.3 | GO:0009597 | detection of virus(GO:0009597) |
| 0.3 | 6.2 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.3 | 3.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.3 | 1.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.3 | 1.8 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.2 | 5.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.2 | 1.0 | GO:0021781 | glial cell fate commitment(GO:0021781) |
| 0.2 | 1.9 | GO:0060025 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) regulation of synaptic activity(GO:0060025) |
| 0.2 | 15.9 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.2 | 1.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.2 | 14.5 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.2 | 2.4 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.2 | 1.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.2 | 1.7 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.2 | 0.8 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.2 | 10.4 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.2 | 1.8 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 1.2 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.2 | 6.3 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.2 | 1.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.2 | 1.5 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.2 | 3.0 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.2 | 0.9 | GO:0032849 | regulation of cellular pH reduction(GO:0032847) positive regulation of cellular pH reduction(GO:0032849) |
| 0.2 | 2.6 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.2 | 0.5 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.2 | 1.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.2 | 4.3 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.2 | 6.7 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 2.6 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.1 | 0.6 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 1.4 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.1 | 5.3 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.1 | 6.8 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 7.1 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.1 | 1.2 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 3.7 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.1 | 0.9 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 0.6 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.1 | 1.9 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 1.9 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 3.9 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 2.8 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 2.6 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 6.7 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 1.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.3 | GO:0098758 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.1 | 12.5 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.1 | 3.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 1.5 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.1 | 1.6 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 11.4 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.1 | 41.3 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.1 | 4.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 0.7 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.1 | 2.1 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.4 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) |
| 0.1 | 2.0 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.5 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 1.0 | GO:0034372 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.1 | 1.0 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 2.1 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.1 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.1 | 0.5 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.3 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.1 | 6.2 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.1 | 2.2 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.1 | 0.5 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.6 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 3.0 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.0 | 3.7 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.1 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.1 | GO:0051821 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) negative regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010768) transmission of virus(GO:0019089) dissemination or transmission of symbiont from host(GO:0044007) dissemination or transmission of organism from other organism involved in symbiotic interaction(GO:0051821) |
| 0.0 | 1.4 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 1.6 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 7.2 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.0 | 0.2 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 3.5 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 2.0 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.5 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 6.4 | GO:0050658 | nucleic acid transport(GO:0050657) RNA transport(GO:0050658) |
| 0.0 | 0.5 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0002865 | negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) |
| 0.0 | 0.4 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.4 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.0 | 1.0 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 1.8 | GO:0071346 | cellular response to interferon-gamma(GO:0071346) |
| 0.0 | 0.8 | GO:0042102 | positive regulation of T cell proliferation(GO:0042102) |
| 0.0 | 0.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.1 | 65.4 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 4.2 | 58.7 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 3.3 | 13.2 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 2.2 | 21.6 | GO:0045180 | basal cortex(GO:0045180) |
| 1.9 | 5.7 | GO:0099569 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) presynaptic cytoskeleton(GO:0099569) |
| 1.7 | 5.0 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 1.6 | 6.3 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 1.3 | 16.5 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 1.2 | 7.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 1.1 | 117.3 | GO:0030315 | T-tubule(GO:0030315) |
| 1.0 | 15.7 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 1.0 | 7.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 1.0 | 20.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 1.0 | 4.9 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.8 | 9.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.7 | 10.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.6 | 3.0 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.6 | 4.8 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.6 | 4.5 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.5 | 2.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.5 | 1.5 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.5 | 7.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.5 | 8.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.5 | 1.8 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.4 | 5.7 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.4 | 11.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.4 | 28.8 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.4 | 5.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.4 | 6.8 | GO:0008091 | spectrin(GO:0008091) |
| 0.4 | 2.0 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.4 | 1.4 | GO:0070695 | FHF complex(GO:0070695) |
| 0.3 | 8.0 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.3 | 3.9 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.3 | 1.9 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.3 | 4.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.3 | 33.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.3 | 8.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.3 | 4.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.3 | 21.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.3 | 5.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.2 | 9.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 12.8 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.2 | 2.7 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 2.7 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 6.2 | GO:0005921 | gap junction(GO:0005921) |
| 0.2 | 53.8 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.2 | 7.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 41.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.2 | 0.8 | GO:0045298 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) tubulin complex(GO:0045298) |
| 0.2 | 12.8 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 4.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 26.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 11.7 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.1 | 3.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 5.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 1.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 3.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 2.0 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 96.7 | GO:0043005 | neuron projection(GO:0043005) |
| 0.1 | 1.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 8.2 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 0.3 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.1 | 8.3 | GO:0098793 | presynapse(GO:0098793) |
| 0.1 | 3.9 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 0.8 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 15.6 | GO:0045121 | membrane raft(GO:0045121) |
| 0.1 | 15.4 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.0 | 0.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 3.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 1.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 2.7 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.0 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 6.9 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.6 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.1 | GO:0044456 | synapse part(GO:0044456) |
| 0.0 | 0.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.9 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.9 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.7 | GO:0015030 | Cajal body(GO:0015030) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.1 | 65.4 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 6.1 | 24.5 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 5.9 | 23.6 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 4.9 | 82.5 | GO:0031432 | titin binding(GO:0031432) |
| 3.2 | 12.6 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 2.6 | 7.7 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 2.4 | 23.7 | GO:0051425 | PTB domain binding(GO:0051425) |
| 2.1 | 16.5 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 1.9 | 33.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 1.7 | 81.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 1.7 | 46.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 1.7 | 28.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 1.4 | 7.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 1.4 | 12.3 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 1.3 | 20.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 1.3 | 10.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 1.2 | 33.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 1.1 | 4.6 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 1.0 | 24.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 1.0 | 4.0 | GO:0003990 | acetylcholinesterase activity(GO:0003990) choline binding(GO:0033265) |
| 1.0 | 4.8 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.9 | 7.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.8 | 12.5 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.8 | 10.4 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.7 | 6.6 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.7 | 7.9 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.5 | 17.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.5 | 1.5 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.5 | 6.7 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.5 | 13.5 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.5 | 28.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.4 | 4.8 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.4 | 1.3 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.4 | 1.7 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.4 | 9.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.4 | 11.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.4 | 2.4 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.4 | 1.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.4 | 1.9 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.4 | 1.9 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.4 | 2.2 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.4 | 21.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.4 | 5.4 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.3 | 10.6 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.3 | 5.7 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.3 | 5.0 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.3 | 0.9 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.3 | 1.8 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.3 | 23.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.3 | 5.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 0.8 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.3 | 2.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 1.3 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.2 | 8.9 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.2 | 1.6 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 3.9 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 3.5 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.2 | 3.5 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.2 | 1.0 | GO:0050610 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.2 | 2.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.2 | 2.3 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.2 | 4.9 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.2 | 1.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 3.0 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 3.7 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.2 | 0.8 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.2 | 1.5 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.2 | 7.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 0.8 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 18.9 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 10.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 4.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.4 | GO:0000035 | acyl binding(GO:0000035) |
| 0.1 | 2.4 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 1.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 17.7 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.1 | 0.8 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 1.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.3 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.1 | 5.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 3.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 3.7 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 1.1 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 12.3 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 11.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 10.0 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 2.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 6.1 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 1.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.6 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 2.3 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 3.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 4.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 1.1 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 15.0 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 1.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 2.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 6.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.2 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.0 | 1.7 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.1 | GO:0004339 | glucan 1,4-alpha-glucosidase activity(GO:0004339) |
| 0.0 | 2.0 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 1.7 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 8.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 9.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 2.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 4.2 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.8 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.3 | GO:0005310 | dicarboxylic acid transmembrane transporter activity(GO:0005310) |
| 0.0 | 0.4 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.1 | GO:0050051 | alkane 1-monooxygenase activity(GO:0018685) leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.0 | 0.7 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 2.0 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 2.2 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 1.1 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 13.6 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 4.8 | GO:0005525 | GTP binding(GO:0005525) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 75.8 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 1.0 | 78.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.8 | 23.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.8 | 34.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.4 | 39.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.4 | 11.6 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.4 | 32.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.3 | 8.5 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.3 | 5.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.3 | 8.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.2 | 12.0 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.2 | 9.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.2 | 6.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 2.7 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 5.0 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 4.8 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 5.1 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 4.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 5.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 2.0 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 8.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 1.9 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 3.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 1.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 0.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 4.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 3.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 9.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.7 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.5 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.2 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.9 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.9 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.3 | PID RAS PATHWAY | Regulation of Ras family activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 65.4 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 1.4 | 37.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 1.2 | 13.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 1.2 | 20.9 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.9 | 12.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.7 | 24.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.6 | 15.5 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.6 | 23.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.5 | 10.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.5 | 100.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.4 | 13.0 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.4 | 9.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.4 | 13.0 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.3 | 11.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.3 | 13.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 5.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 7.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 5.1 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.2 | 4.0 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 19.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 5.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.2 | 7.1 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.2 | 6.7 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 0.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 8.5 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 3.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 4.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 7.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.8 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 3.0 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 4.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.5 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 1.7 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 4.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.1 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 6.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 3.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 1.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 1.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 0.7 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.1 | 0.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.0 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 4.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 3.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.4 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.2 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.1 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |