Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for NR4A3
Z-value: 0.79
Transcription factors associated with NR4A3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR4A3
|
ENSG00000119508.18 | NR4A3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR4A3 | hg38_v1_chr9_+_99821846_99821862 | -0.25 | 1.9e-04 | Click! |
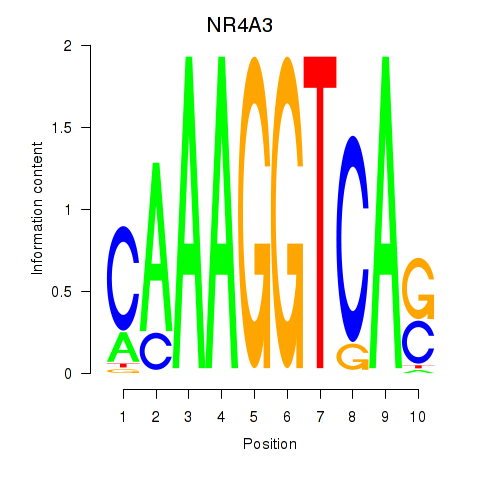
Activity profile of NR4A3 motif
Sorted Z-values of NR4A3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NR4A3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 19.6 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 2.2 | 6.7 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 2.0 | 6.0 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 1.9 | 5.6 | GO:0060623 | regulation of chromosome condensation(GO:0060623) cellular response to iron(III) ion(GO:0071283) |
| 1.9 | 24.2 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 1.8 | 7.3 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 1.8 | 5.4 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 1.7 | 5.2 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 1.7 | 5.1 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 1.7 | 6.7 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 1.5 | 4.6 | GO:2000276 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 1.4 | 4.1 | GO:0060164 | midbrain-hindbrain boundary morphogenesis(GO:0021555) trochlear nerve development(GO:0021558) regulation of timing of neuron differentiation(GO:0060164) |
| 1.3 | 3.9 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 1.3 | 12.8 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 1.2 | 7.3 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 1.2 | 4.7 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 1.1 | 3.3 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 1.1 | 11.9 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 1.1 | 3.2 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 1.0 | 9.0 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 1.0 | 5.8 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 1.0 | 2.9 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 1.0 | 2.9 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.9 | 2.7 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) phenylpropanoid catabolic process(GO:0046271) |
| 0.9 | 2.7 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 0.9 | 2.6 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.9 | 2.6 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.8 | 0.8 | GO:0036115 | fatty-acyl-CoA catabolic process(GO:0036115) |
| 0.8 | 4.8 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.8 | 6.3 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.8 | 2.3 | GO:0060957 | endocardial cell fate commitment(GO:0060957) endocardial cushion cell fate commitment(GO:0061445) |
| 0.8 | 3.0 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.7 | 10.5 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.7 | 3.6 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.7 | 2.8 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.7 | 2.8 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.7 | 5.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.7 | 3.3 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.6 | 3.7 | GO:0003164 | His-Purkinje system development(GO:0003164) |
| 0.6 | 0.6 | GO:0050787 | detoxification of mercury ion(GO:0050787) |
| 0.6 | 2.4 | GO:1904116 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.6 | 1.8 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.6 | 9.5 | GO:1900364 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.6 | 2.4 | GO:0046833 | snRNA export from nucleus(GO:0006408) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.6 | 2.3 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.6 | 1.7 | GO:0070343 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.6 | 6.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.5 | 2.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.5 | 2.6 | GO:1901162 | spermidine biosynthetic process(GO:0008295) primary amino compound biosynthetic process(GO:1901162) |
| 0.5 | 7.0 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.5 | 6.0 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.5 | 1.5 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.5 | 2.9 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.5 | 4.7 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.5 | 1.4 | GO:0021539 | subthalamus development(GO:0021539) subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) left lung development(GO:0060459) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.5 | 11.3 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.5 | 1.8 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.4 | 2.7 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.4 | 3.5 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.4 | 1.8 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.4 | 7.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.4 | 1.7 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.4 | 8.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.4 | 2.6 | GO:0031337 | cellular response to phosphate starvation(GO:0016036) positive regulation of sulfur amino acid metabolic process(GO:0031337) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.4 | 1.3 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.4 | 28.0 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.4 | 2.0 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.4 | 1.2 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.4 | 5.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.4 | 2.3 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.4 | 1.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.4 | 28.9 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.4 | 1.1 | GO:0055018 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm development(GO:0048389) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) pattern specification involved in mesonephros development(GO:0061227) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in kidney development(GO:0072098) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.4 | 4.8 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.4 | 1.8 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 0.4 | 1.1 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.4 | 1.8 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.3 | 1.4 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.3 | 2.4 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.3 | 1.7 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.3 | 2.7 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.3 | 1.0 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.3 | 4.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.3 | 1.8 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.3 | 4.9 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.3 | 1.8 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.3 | 1.4 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.3 | 1.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.3 | 1.9 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.3 | 1.1 | GO:1900104 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.3 | 4.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.3 | 1.1 | GO:0032072 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.3 | 1.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.3 | 3.2 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.3 | 5.0 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.3 | 1.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.3 | 1.8 | GO:1904431 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) positive regulation of t-circle formation(GO:1904431) |
| 0.3 | 2.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.3 | 3.3 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.2 | 2.0 | GO:0044206 | pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) |
| 0.2 | 1.2 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.2 | 0.9 | GO:0003290 | septum secundum development(GO:0003285) atrial septum secundum morphogenesis(GO:0003290) embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.2 | 0.9 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.2 | 6.8 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.2 | 0.9 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.2 | 1.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.2 | 1.1 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.2 | 0.9 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.2 | 1.1 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.2 | 0.7 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.2 | 0.7 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.2 | 0.7 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.2 | 2.4 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.2 | 13.2 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.2 | 1.7 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.2 | 11.4 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.2 | 2.4 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.2 | 2.0 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.2 | 1.8 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.2 | 0.6 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.2 | 8.1 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.2 | 3.2 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.2 | 0.6 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.2 | 4.3 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.2 | 7.3 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.2 | 2.8 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.2 | 1.0 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.2 | 2.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 0.3 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.2 | 1.4 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.2 | 2.0 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.2 | 0.8 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.2 | 1.0 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.2 | 3.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 | 0.8 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.2 | 3.6 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.2 | 0.5 | GO:0032912 | negative regulation of transforming growth factor beta2 production(GO:0032912) negative regulation of hair follicle maturation(GO:0048817) regulation of melanosome transport(GO:1902908) |
| 0.2 | 4.3 | GO:0007567 | parturition(GO:0007567) |
| 0.2 | 1.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.1 | 0.9 | GO:0051563 | microglia differentiation(GO:0014004) microglia development(GO:0014005) collateral sprouting in absence of injury(GO:0048669) smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) response to norepinephrine(GO:0071873) cellular response to norepinephrine stimulus(GO:0071874) |
| 0.1 | 0.6 | GO:0032764 | smooth muscle hyperplasia(GO:0014806) negative regulation of mast cell cytokine production(GO:0032764) |
| 0.1 | 1.5 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 1.6 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 1.5 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.1 | 0.6 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.1 | 1.6 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 7.6 | GO:0006735 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 3.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.5 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.1 | 0.9 | GO:0010269 | response to selenium ion(GO:0010269) negative regulation of serotonin secretion(GO:0014063) |
| 0.1 | 2.7 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 2.9 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.1 | 0.5 | GO:0019376 | galactolipid catabolic process(GO:0019376) |
| 0.1 | 0.8 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 3.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 2.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.8 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 0.5 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.1 | 1.3 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 7.8 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.4 | GO:0072141 | renal interstitial fibroblast development(GO:0072141) |
| 0.1 | 3.2 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 1.6 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 0.8 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 3.4 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 3.7 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.1 | 3.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.5 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 1.0 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.1 | 0.4 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.1 | 0.3 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.1 | 1.6 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.1 | 2.0 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 0.9 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 0.5 | GO:1901523 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.1 | 2.7 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.1 | 2.0 | GO:2000404 | regulation of T cell migration(GO:2000404) |
| 0.1 | 0.2 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.1 | 0.5 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 1.3 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.1 | 2.9 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 0.9 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 0.8 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 0.2 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.1 | 2.8 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 1.3 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 3.2 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 0.9 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.1 | 1.0 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 2.2 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 1.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 5.7 | GO:0051893 | regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.1 | 0.2 | GO:0048865 | stem cell fate commitment(GO:0048865) |
| 0.1 | 1.6 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.1 | 0.3 | GO:0036109 | alpha-linolenic acid metabolic process(GO:0036109) |
| 0.1 | 1.1 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 0.1 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.1 | 1.4 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.1 | 7.1 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.1 | 1.5 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.1 | 2.4 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 0.8 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 1.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.2 | GO:0030167 | proteoglycan catabolic process(GO:0030167) regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.6 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 1.8 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 8.4 | GO:0007565 | female pregnancy(GO:0007565) |
| 0.0 | 3.0 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 4.0 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.5 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.8 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 1.2 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 1.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 1.1 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 1.1 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.4 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 1.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.6 | GO:1901798 | positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 1.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.3 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 1.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.6 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.1 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.8 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.0 | 0.4 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 0.2 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.4 | GO:0010744 | positive regulation of macrophage derived foam cell differentiation(GO:0010744) |
| 0.0 | 1.5 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 3.9 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.0 | 0.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.5 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.5 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 2.3 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 0.1 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.1 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.0 | 0.1 | GO:1990822 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.8 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.8 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.7 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.8 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 3.4 | GO:0043687 | post-translational protein modification(GO:0043687) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 24.2 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 2.0 | 26.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 1.7 | 14.9 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 1.7 | 8.3 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 1.5 | 9.0 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 1.4 | 17.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 1.4 | 5.6 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 1.4 | 5.5 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 1.4 | 9.5 | GO:0032021 | NELF complex(GO:0032021) |
| 1.2 | 3.6 | GO:0036029 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 1.0 | 11.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.8 | 4.2 | GO:0001652 | granular component(GO:0001652) |
| 0.8 | 2.4 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.8 | 6.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.8 | 7.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.8 | 2.3 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.7 | 2.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.6 | 1.9 | GO:0071750 | dimeric IgA immunoglobulin complex(GO:0071750) secretory dimeric IgA immunoglobulin complex(GO:0071752) |
| 0.6 | 1.8 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.6 | 2.4 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.6 | 2.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.5 | 5.8 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.5 | 6.7 | GO:0030677 | ribonuclease P complex(GO:0030677) |
| 0.5 | 20.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.5 | 3.6 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.5 | 3.3 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.5 | 2.7 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.4 | 28.0 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.4 | 3.0 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.4 | 2.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.4 | 2.7 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.3 | 2.4 | GO:0016589 | NURF complex(GO:0016589) |
| 0.3 | 2.4 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.3 | 1.0 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.3 | 1.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.3 | 6.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.3 | 8.2 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.3 | 16.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.3 | 3.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 3.8 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.2 | 1.8 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.2 | 0.7 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.2 | 3.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.2 | 2.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 4.6 | GO:0016469 | proton-transporting two-sector ATPase complex(GO:0016469) |
| 0.2 | 0.7 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.2 | 1.3 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.2 | 1.5 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.2 | 3.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.2 | 1.6 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.2 | 7.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 0.8 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.1 | 1.3 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 3.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 2.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 1.3 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 3.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.0 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 1.5 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 0.9 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 5.6 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 1.4 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 2.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 1.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 1.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 2.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 8.3 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.1 | 2.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 3.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 17.0 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 2.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.8 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 1.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 3.9 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 0.5 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.1 | 2.7 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 1.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 3.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 1.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 0.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.3 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 0.9 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 1.8 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 17.4 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.1 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 2.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 5.4 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 2.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 2.3 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 1.1 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 5.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.1 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 1.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 2.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 1.7 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 3.5 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 1.7 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 1.1 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 2.8 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 1.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.9 | GO:0036064 | ciliary basal body(GO:0036064) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.3 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 1.7 | 5.1 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 1.7 | 6.7 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 1.4 | 5.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 1.3 | 10.5 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 1.2 | 7.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 1.2 | 7.2 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 1.2 | 11.9 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 1.1 | 3.3 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 1.1 | 7.5 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.9 | 2.8 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.9 | 5.6 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.9 | 7.3 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.9 | 2.7 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.8 | 6.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.7 | 11.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.7 | 2.8 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.7 | 2.7 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.7 | 24.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.6 | 5.8 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.6 | 1.8 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.6 | 2.3 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.6 | 5.5 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.5 | 2.6 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.5 | 28.0 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.5 | 1.5 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.5 | 1.5 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.5 | 2.5 | GO:0019862 | IgA binding(GO:0019862) |
| 0.5 | 2.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.5 | 2.9 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.5 | 6.7 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.5 | 3.7 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.4 | 5.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.4 | 2.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.4 | 11.2 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.4 | 3.6 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.4 | 2.3 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.4 | 1.8 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.4 | 1.5 | GO:0031716 | calcitonin receptor activity(GO:0004948) calcitonin receptor binding(GO:0031716) |
| 0.4 | 1.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.3 | 1.4 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.3 | 5.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.3 | 9.9 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.3 | 6.0 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.3 | 3.2 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.3 | 0.9 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.3 | 1.8 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.3 | 2.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.3 | 2.0 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.3 | 1.7 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) 5'-flap endonuclease activity(GO:0017108) |
| 0.3 | 2.6 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.3 | 2.5 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.3 | 1.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.3 | 1.1 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.3 | 1.4 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.3 | 6.6 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.3 | 1.3 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.3 | 2.0 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.3 | 4.6 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 2.4 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.2 | 1.2 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.2 | 6.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 3.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.2 | 2.4 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.2 | 4.8 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.2 | 1.1 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.2 | 0.8 | GO:0004341 | gluconolactonase activity(GO:0004341) |
| 0.2 | 1.0 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.2 | 3.0 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.2 | 1.8 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.2 | 1.5 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 4.9 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.2 | 1.3 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.2 | 1.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 3.6 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.2 | 0.9 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.2 | 1.6 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.2 | 12.4 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.2 | 2.6 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.2 | 2.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 1.2 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.2 | 1.7 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 9.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 1.3 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.2 | 0.5 | GO:0097258 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.2 | 0.7 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.2 | 0.7 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.2 | 3.0 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.2 | 4.7 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.2 | 2.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 1.2 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.2 | 1.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.2 | 2.4 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 2.0 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.4 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 3.5 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 3.5 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 14.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 2.7 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.1 | 0.8 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.6 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 1.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.4 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 0.6 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 1.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.6 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 2.0 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 3.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 23.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 2.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 3.0 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.7 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.9 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.1 | 0.6 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.1 | 1.9 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.8 | GO:0038064 | collagen receptor activity(GO:0038064) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 4.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.8 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.1 | 5.8 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.1 | 3.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 5.1 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 11.0 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 3.5 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 4.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 2.3 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 1.1 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 3.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.2 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 2.1 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 2.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 5.4 | GO:0070405 | ammonium ion binding(GO:0070405) |
| 0.1 | 2.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.8 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.6 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 0.3 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.1 | 0.3 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.3 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.1 | 0.2 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.1 | 1.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 1.8 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 4.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 2.0 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 1.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 1.5 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.5 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.3 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.5 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.6 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 2.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.7 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 2.3 | GO:0032451 | demethylase activity(GO:0032451) |
| 0.0 | 3.1 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 5.4 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 1.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 3.2 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 1.1 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 1.7 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 1.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.8 | GO:0051429 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 2.6 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 0.9 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.6 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 1.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 1.0 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.8 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 1.1 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 3.0 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 1.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 24.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 2.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 13.0 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 4.5 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 5.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 31.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 3.8 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 6.2 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 1.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 0.8 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 1.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 6.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 0.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 1.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 1.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 6.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 1.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 5.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.8 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 2.8 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 2.8 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.1 | 1.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 3.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 0.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 0.4 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 3.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 4.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.9 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 3.2 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.5 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 2.9 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 3.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.1 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.9 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 1.0 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 2.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 5.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 25.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.8 | 70.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.5 | 15.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.4 | 5.6 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.4 | 6.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.4 | 8.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.3 | 3.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.3 | 10.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.3 | 18.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.3 | 5.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.3 | 2.6 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.3 | 2.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 3.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.2 | 6.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 2.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 4.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 6.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.2 | 3.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.2 | 3.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 15.6 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.1 | 1.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 3.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 4.5 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 4.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.5 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 3.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.8 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 1.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 2.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.9 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 7.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.7 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 3.8 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 1.7 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 2.0 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 3.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 0.9 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 2.1 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 1.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 0.8 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 2.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 1.7 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 2.0 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 0.9 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 5.7 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.1 | 6.0 | REACTOME HOST INTERACTIONS OF HIV FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.1 | 2.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 2.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 4.0 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 2.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 3.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 5.0 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 2.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 2.7 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 1.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 6.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.4 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 3.7 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 1.1 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.0 | 0.9 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.4 | REACTOME TRIF MEDIATED TLR3 SIGNALING | Genes involved in TRIF mediated TLR3 signaling |
| 0.0 | 0.7 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 1.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 6.3 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 5.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 3.3 | REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | Genes involved in MyD88:Mal cascade initiated on plasma membrane |
| 0.0 | 1.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.5 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |