Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for PBX2
Z-value: 0.32
Transcription factors associated with PBX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
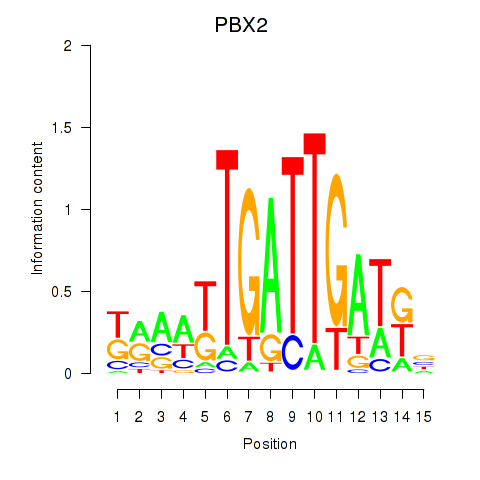
PBX2
|
ENSG00000204304.12 | PBX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PBX2 | hg38_v1_chr6_-_32190170_32190214 | -0.12 | 7.4e-02 | Click! |
Activity profile of PBX2 motif
Sorted Z-values of PBX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PBX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_120604199 | 4.33 |
ENST00000371315.3
|
MCTS1
|
MCTS1 re-initiation and release factor |
| chrX_+_120604084 | 4.01 |
ENST00000371317.10
|
MCTS1
|
MCTS1 re-initiation and release factor |
| chr12_+_53441724 | 2.93 |
ENST00000551003.5
ENST00000429243.7 ENST00000549068.5 ENST00000549740.5 ENST00000546581.5 ENST00000549581.5 ENST00000547368.5 ENST00000379786.8 ENST00000551945.5 ENST00000547717.1 |
PRR13
ENSG00000257379.1
|
proline rich 13 novel transcript |
| chr7_+_142791635 | 2.73 |
ENST00000633705.1
|
TRBC1
|
T cell receptor beta constant 1 |
| chrX_+_56563569 | 2.69 |
ENST00000338222.7
|
UBQLN2
|
ubiquilin 2 |
| chr14_-_55191534 | 2.48 |
ENST00000395425.6
ENST00000247191.7 |
DLGAP5
|
DLG associated protein 5 |
| chr2_+_233693659 | 2.28 |
ENST00000406651.1
|
UGT1A6
|
UDP glucuronosyltransferase family 1 member A6 |
| chr3_-_129440007 | 2.00 |
ENST00000503197.5
ENST00000249910.5 ENST00000507208.1 ENST00000393278.6 |
MBD4
|
methyl-CpG binding domain 4, DNA glycosylase |
| chr2_+_233692881 | 1.95 |
ENST00000305139.11
|
UGT1A6
|
UDP glucuronosyltransferase family 1 member A6 |
| chr14_+_64388296 | 1.92 |
ENST00000554739.5
ENST00000554768.6 ENST00000652179.1 ENST00000652337.1 ENST00000557370.3 |
MTHFD1
|
methylenetetrahydrofolate dehydrogenase, cyclohydrolase and formyltetrahydrofolate synthetase 1 |
| chr3_-_129439925 | 1.91 |
ENST00000429544.7
|
MBD4
|
methyl-CpG binding domain 4, DNA glycosylase |
| chr1_+_101237009 | 1.90 |
ENST00000305352.7
|
S1PR1
|
sphingosine-1-phosphate receptor 1 |
| chr2_-_264024 | 1.86 |
ENST00000403712.6
ENST00000356150.10 ENST00000626873.2 ENST00000405430.5 |
SH3YL1
|
SH3 and SYLF domain containing 1 |
| chr2_+_12716893 | 1.84 |
ENST00000381465.2
ENST00000155926.9 |
TRIB2
|
tribbles pseudokinase 2 |
| chr12_-_27972725 | 1.70 |
ENST00000545234.6
|
PTHLH
|
parathyroid hormone like hormone |
| chr12_-_27971970 | 1.60 |
ENST00000395872.5
ENST00000201015.8 |
PTHLH
|
parathyroid hormone like hormone |
| chr22_-_30246739 | 1.49 |
ENST00000403987.3
ENST00000249075.4 |
LIF
|
LIF interleukin 6 family cytokine |
| chr17_-_30824665 | 1.45 |
ENST00000324238.7
|
CRLF3
|
cytokine receptor like factor 3 |
| chr3_+_190615308 | 1.40 |
ENST00000412080.1
|
IL1RAP
|
interleukin 1 receptor accessory protein |
| chr11_-_62665169 | 1.38 |
ENST00000528405.1
ENST00000525675.1 ENST00000524958.6 |
ENSG00000255432.1
C11orf98
|
novel protein chromosome 11 open reading frame 98 |
| chr2_+_20447065 | 1.15 |
ENST00000272233.6
|
RHOB
|
ras homolog family member B |
| chrM_+_12329 | 1.00 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 5 |
| chr7_-_32490361 | 0.95 |
ENST00000410044.5
ENST00000450169.7 ENST00000409987.5 ENST00000409782.5 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr7_-_151520080 | 0.80 |
ENST00000496004.5
|
RHEB
|
Ras homolog, mTORC1 binding |
| chr6_+_159790448 | 0.78 |
ENST00000367034.5
|
MRPL18
|
mitochondrial ribosomal protein L18 |
| chr2_+_44168866 | 0.78 |
ENST00000282412.9
ENST00000409432.7 ENST00000378551.6 ENST00000345249.8 |
PPM1B
|
protein phosphatase, Mg2+/Mn2+ dependent 1B |
| chr10_+_122560751 | 0.74 |
ENST00000338354.10
ENST00000664692.1 ENST00000653442.1 ENST00000664974.1 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr10_+_122560639 | 0.69 |
ENST00000344338.7
ENST00000330163.8 ENST00000652446.2 ENST00000666315.1 ENST00000368955.7 ENST00000368909.7 ENST00000368956.6 ENST00000619379.1 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr10_+_122560679 | 0.68 |
ENST00000657942.1
|
DMBT1
|
deleted in malignant brain tumors 1 |
| chr14_-_23155302 | 0.60 |
ENST00000529705.6
|
SLC7A8
|
solute carrier family 7 member 8 |
| chr17_+_8288637 | 0.56 |
ENST00000407006.8
ENST00000226105.11 ENST00000580434.5 ENST00000439238.3 |
RANGRF
|
RAN guanine nucleotide release factor |
| chr15_+_67125707 | 0.48 |
ENST00000540846.6
|
SMAD3
|
SMAD family member 3 |
| chr5_+_55024250 | 0.42 |
ENST00000231009.3
|
GZMK
|
granzyme K |
| chr14_+_22516273 | 0.41 |
ENST00000390510.1
|
TRAJ27
|
T cell receptor alpha joining 27 |
| chr8_-_6563409 | 0.37 |
ENST00000325203.9
|
ANGPT2
|
angiopoietin 2 |
| chr2_+_203867943 | 0.33 |
ENST00000295854.10
ENST00000487393.1 ENST00000472206.1 |
CTLA4
|
cytotoxic T-lymphocyte associated protein 4 |
| chr11_-_55936400 | 0.28 |
ENST00000301532.3
|
OR5I1
|
olfactory receptor family 5 subfamily I member 1 |
| chr4_+_37960397 | 0.26 |
ENST00000504686.2
|
PTTG2
|
pituitary tumor-transforming 2 |
| chrX_+_46837034 | 0.25 |
ENST00000218340.4
|
RP2
|
RP2 activator of ARL3 GTPase |
| chr7_-_113087720 | 0.25 |
ENST00000297146.7
|
GPR85
|
G protein-coupled receptor 85 |
| chr17_-_1229706 | 0.23 |
ENST00000574139.7
|
ABR
|
ABR activator of RhoGEF and GTPase |
| chr8_+_109086585 | 0.21 |
ENST00000518632.2
|
TRHR
|
thyrotropin releasing hormone receptor |
| chr2_+_203867764 | 0.20 |
ENST00000648405.2
|
CTLA4
|
cytotoxic T-lymphocyte associated protein 4 |
| chr16_+_1078781 | 0.12 |
ENST00000293897.5
|
SSTR5
|
somatostatin receptor 5 |
| chr12_+_41437680 | 0.10 |
ENST00000649474.1
ENST00000539469.6 ENST00000298919.7 |
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr14_+_58298497 | 0.09 |
ENST00000348476.7
ENST00000355431.8 ENST00000395168.7 |
ARID4A
|
AT-rich interaction domain 4A |
| chrX_+_1615049 | 0.09 |
ENST00000381241.9
|
ASMT
|
acetylserotonin O-methyltransferase |
| chr3_-_52830664 | 0.06 |
ENST00000266041.9
ENST00000406595.5 ENST00000485816.5 |
ITIH4
|
inter-alpha-trypsin inhibitor heavy chain 4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.3 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.7 | 2.7 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of G-protein coupled receptor internalization(GO:1904020) negative regulation of G-protein coupled receptor internalization(GO:1904021) |
| 0.6 | 1.9 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.5 | 1.5 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.5 | 1.9 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.5 | 1.8 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.4 | 2.5 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.4 | 4.2 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.3 | 3.9 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.3 | 0.8 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.2 | 1.4 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.2 | 2.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 0.6 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.1 | 0.4 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.1 | 0.8 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.1 | 3.3 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 0.5 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.1 | 0.5 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 0.8 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.1 | 0.2 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 1.9 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.0 | 1.1 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 2.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0097368 | histone H4-K20 trimethylation(GO:0034773) establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.1 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 1.0 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.6 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 1.4 | GO:0046427 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.0 | 1.0 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 2.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 8.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 2.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 0.3 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.1 | 0.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 2.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 2.7 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.1 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.0 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.5 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.9 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.7 | 2.1 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.5 | 1.9 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.2 | 1.4 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.2 | 1.9 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.1 | 1.5 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 8.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 1.8 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 4.2 | GO:0015020 | retinoic acid binding(GO:0001972) glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.5 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 3.3 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.6 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 0.8 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 2.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.6 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 1.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.0 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.8 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.9 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 2.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.1 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.9 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.2 | 4.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.0 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.8 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 3.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.5 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |