|
chr8_-_79767843
Show fit
|
11.77 |
ENST00000337919.9
|
HEY1
|
hes related family bHLH transcription factor with YRPW motif 1
|
|
chr17_-_58529277
Show fit
|
11.41 |
ENST00000579371.5
|
SEPTIN4
|
septin 4
|
|
chr17_-_58529344
Show fit
|
11.38 |
ENST00000317268.7
|
SEPTIN4
|
septin 4
|
|
chr17_-_58529303
Show fit
|
11.13 |
ENST00000580844.5
|
SEPTIN4
|
septin 4
|
|
chr8_-_79767462
Show fit
|
10.46 |
ENST00000674295.1
ENST00000518733.1
ENST00000674418.1
ENST00000674358.1
ENST00000354724.8
|
HEY1
|
hes related family bHLH transcription factor with YRPW motif 1
|
|
chr4_-_175812746
Show fit
|
9.02 |
ENST00000393658.6
|
GPM6A
|
glycoprotein M6A
|
|
chr4_+_155667654
Show fit
|
8.86 |
ENST00000513574.1
|
GUCY1A1
|
guanylate cyclase 1 soluble subunit alpha 1
|
|
chr9_-_137028878
Show fit
|
8.42 |
ENST00000625103.1
ENST00000614293.4
|
ABCA2
|
ATP binding cassette subfamily A member 2
|
|
chr12_+_12785652
Show fit
|
8.02 |
ENST00000356591.5
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr12_-_16606795
Show fit
|
7.63 |
ENST00000447609.5
|
LMO3
|
LIM domain only 3
|
|
chr3_+_142723999
Show fit
|
7.59 |
ENST00000476941.6
ENST00000273482.10
|
TRPC1
|
transient receptor potential cation channel subfamily C member 1
|
|
chr8_-_22232020
Show fit
|
7.28 |
ENST00000454243.7
ENST00000321613.7
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein
|
|
chr16_-_70685975
Show fit
|
7.22 |
ENST00000338779.11
|
MTSS2
|
MTSS I-BAR domain containing 2
|
|
chr10_-_37857582
Show fit
|
6.92 |
ENST00000395867.8
ENST00000611278.4
|
ZNF248
|
zinc finger protein 248
|
|
chr4_-_86594037
Show fit
|
6.91 |
ENST00000641050.1
ENST00000641831.1
ENST00000515400.3
ENST00000641391.1
ENST00000641157.1
ENST00000641737.1
ENST00000502302.6
ENST00000640527.1
ENST00000512046.2
ENST00000513186.7
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr4_+_155667198
Show fit
|
6.77 |
ENST00000296518.11
|
GUCY1A1
|
guanylate cyclase 1 soluble subunit alpha 1
|
|
chr19_+_50203607
Show fit
|
6.60 |
ENST00000642316.2
ENST00000425460.6
ENST00000440075.6
ENST00000376970.6
ENST00000599920.5
|
MYH14
|
myosin heavy chain 14
|
|
chr10_-_37858037
Show fit
|
6.06 |
ENST00000395873.7
ENST00000357328.8
ENST00000395874.2
|
ZNF248
|
zinc finger protein 248
|
|
chr6_+_30884353
Show fit
|
6.02 |
ENST00000428153.6
ENST00000376568.8
ENST00000452441.5
ENST00000515219.5
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr20_-_49278416
Show fit
|
5.89 |
ENST00000371754.8
|
ZNFX1
|
zinc finger NFX1-type containing 1
|
|
chr4_+_155667096
Show fit
|
5.73 |
ENST00000393832.7
|
GUCY1A1
|
guanylate cyclase 1 soluble subunit alpha 1
|
|
chr4_-_73069696
Show fit
|
5.71 |
ENST00000507544.3
ENST00000295890.8
|
COX18
|
cytochrome c oxidase assembly factor COX18
|
|
chr17_-_35930727
Show fit
|
5.29 |
ENST00000616596.4
ENST00000612980.4
ENST00000613308.4
ENST00000619876.4
|
RDM1
|
RAD52 motif containing 1
|
|
chr19_-_38899800
Show fit
|
5.28 |
ENST00000414941.5
ENST00000358931.9
ENST00000392081.6
|
SIRT2
|
sirtuin 2
|
|
chr5_-_140564245
Show fit
|
5.23 |
ENST00000412920.7
ENST00000511201.2
ENST00000354402.9
ENST00000356738.6
|
APBB3
|
amyloid beta precursor protein binding family B member 3
|
|
chr6_+_30884063
Show fit
|
4.99 |
ENST00000511510.5
ENST00000376569.7
ENST00000376570.8
ENST00000504927.5
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr17_+_2796404
Show fit
|
4.98 |
ENST00000366401.8
ENST00000254695.13
ENST00000542807.1
|
RAP1GAP2
|
RAP1 GTPase activating protein 2
|
|
chr8_-_126557691
Show fit
|
4.86 |
ENST00000652209.1
|
LRATD2
|
LRAT domain containing 2
|
|
chr1_-_223363337
Show fit
|
4.78 |
ENST00000608996.5
|
SUSD4
|
sushi domain containing 4
|
|
chr2_+_218959635
Show fit
|
4.72 |
ENST00000302625.6
|
CDK5R2
|
cyclin dependent kinase 5 regulatory subunit 2
|
|
chr3_+_159852933
Show fit
|
4.61 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr1_+_14945775
Show fit
|
4.52 |
ENST00000400797.3
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr20_+_44582549
Show fit
|
4.52 |
ENST00000372886.6
|
PKIG
|
cAMP-dependent protein kinase inhibitor gamma
|
|
chr11_+_112961480
Show fit
|
4.44 |
ENST00000621850.4
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr14_-_52791597
Show fit
|
4.39 |
ENST00000216410.8
ENST00000557604.1
|
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1
|
|
chr19_-_38899529
Show fit
|
4.34 |
ENST00000249396.12
ENST00000437828.5
|
SIRT2
|
sirtuin 2
|
|
chr19_-_42242526
Show fit
|
4.31 |
ENST00000222330.8
ENST00000676537.1
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr20_-_46308485
Show fit
|
4.30 |
ENST00000537909.4
|
CDH22
|
cadherin 22
|
|
chr11_+_112961402
Show fit
|
4.25 |
ENST00000613217.4
ENST00000316851.12
ENST00000620046.4
ENST00000531044.5
ENST00000529356.5
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr16_+_1612337
Show fit
|
4.24 |
ENST00000674071.1
ENST00000397412.8
|
CRAMP1
|
cramped chromatin regulator homolog 1
|
|
chr1_-_223364059
Show fit
|
4.24 |
ENST00000343846.7
ENST00000484758.6
ENST00000344029.6
ENST00000366878.9
ENST00000494793.6
ENST00000681285.1
ENST00000680429.1
ENST00000681669.1
ENST00000681305.1
|
SUSD4
|
sushi domain containing 4
|
|
chr16_-_30091226
Show fit
|
4.17 |
ENST00000279386.6
ENST00000627355.2
|
TBX6
|
T-box transcription factor 6
|
|
chr6_+_42564060
Show fit
|
4.11 |
ENST00000372903.6
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2
|
|
chr1_+_36224410
Show fit
|
4.09 |
ENST00000469141.6
ENST00000648638.1
ENST00000354618.10
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr11_+_112961247
Show fit
|
3.99 |
ENST00000621518.4
ENST00000618266.4
ENST00000615112.4
ENST00000615285.4
ENST00000619839.4
ENST00000401611.6
ENST00000621128.4
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr11_-_31817904
Show fit
|
3.96 |
ENST00000423822.7
|
PAX6
|
paired box 6
|
|
chr11_-_31817937
Show fit
|
3.92 |
ENST00000638965.1
ENST00000241001.13
ENST00000638914.3
ENST00000379132.8
ENST00000379129.7
|
PAX6
|
paired box 6
|
|
chr19_-_45792755
Show fit
|
3.91 |
ENST00000377735.7
ENST00000270223.7
|
DMWD
|
DM1 locus, WD repeat containing
|
|
chr19_-_55179390
Show fit
|
3.77 |
ENST00000590851.5
|
SYT5
|
synaptotagmin 5
|
|
chr16_-_1611985
Show fit
|
3.63 |
ENST00000426508.7
|
IFT140
|
intraflagellar transport 140
|
|
chr14_-_52791462
Show fit
|
3.60 |
ENST00000650397.1
ENST00000554230.5
|
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1
|
|
chr11_+_66744618
Show fit
|
3.60 |
ENST00000524551.5
ENST00000525908.6
ENST00000540737.7
ENST00000527634.5
|
C11orf80
|
chromosome 11 open reading frame 80
|
|
chrX_-_153971169
Show fit
|
3.53 |
ENST00000369984.4
|
HCFC1
|
host cell factor C1
|
|
chr10_-_97445850
Show fit
|
3.51 |
ENST00000477692.6
ENST00000485122.6
ENST00000370886.9
ENST00000370885.8
ENST00000370902.8
ENST00000370884.5
|
EXOSC1
|
exosome component 1
|
|
chr1_+_50106265
Show fit
|
3.40 |
ENST00000357083.8
|
ELAVL4
|
ELAV like RNA binding protein 4
|
|
chr12_-_48865863
Show fit
|
3.34 |
ENST00000309739.6
|
RND1
|
Rho family GTPase 1
|
|
chr21_+_6499203
Show fit
|
3.32 |
ENST00000619537.5
|
CRYAA2
|
crystallin alpha A2
|
|
chr10_+_116545907
Show fit
|
3.32 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase
|
|
chr7_-_120858066
Show fit
|
3.16 |
ENST00000222747.8
|
TSPAN12
|
tetraspanin 12
|
|
chr21_-_31558977
Show fit
|
3.12 |
ENST00000286827.7
ENST00000541036.5
|
TIAM1
|
TIAM Rac1 associated GEF 1
|
|
chr5_-_116536458
Show fit
|
3.05 |
ENST00000510263.5
|
SEMA6A
|
semaphorin 6A
|
|
chr11_+_66744831
Show fit
|
2.99 |
ENST00000642265.1
ENST00000532565.6
|
C11orf80
|
chromosome 11 open reading frame 80
|
|
chr7_+_74209947
Show fit
|
2.91 |
ENST00000475494.5
ENST00000398475.5
|
LAT2
|
linker for activation of T cells family member 2
|
|
chrX_+_16123544
Show fit
|
2.88 |
ENST00000380289.3
|
GRPR
|
gastrin releasing peptide receptor
|
|
chr16_-_10182394
Show fit
|
2.84 |
ENST00000330684.4
|
GRIN2A
|
glutamate ionotropic receptor NMDA type subunit 2A
|
|
chrX_+_123859807
Show fit
|
2.82 |
ENST00000434753.7
ENST00000430625.1
|
XIAP
|
X-linked inhibitor of apoptosis
|
|
chr10_+_111077021
Show fit
|
2.81 |
ENST00000280155.4
|
ADRA2A
|
adrenoceptor alpha 2A
|
|
chr10_+_73168111
Show fit
|
2.79 |
ENST00000242505.11
|
FAM149B1
|
family with sequence similarity 149 member B1
|
|
chr11_-_32430811
Show fit
|
2.77 |
ENST00000379079.8
ENST00000530998.5
|
WT1
|
WT1 transcription factor
|
|
chr9_-_127950716
Show fit
|
2.70 |
ENST00000373084.8
|
FAM102A
|
family with sequence similarity 102 member A
|
|
chr13_-_24922788
Show fit
|
2.66 |
ENST00000381884.9
|
CENPJ
|
centromere protein J
|
|
chr11_+_118607598
Show fit
|
2.66 |
ENST00000600882.6
ENST00000356063.9
|
PHLDB1
|
pleckstrin homology like domain family B member 1
|
|
chr9_+_99821876
Show fit
|
2.62 |
ENST00000395097.7
|
NR4A3
|
nuclear receptor subfamily 4 group A member 3
|
|
chr14_-_21526312
Show fit
|
2.54 |
ENST00000537235.2
|
SALL2
|
spalt like transcription factor 2
|
|
chr3_-_184017863
Show fit
|
2.54 |
ENST00000427120.6
ENST00000334444.11
ENST00000392579.6
ENST00000382494.6
ENST00000265586.10
ENST00000446941.2
|
ABCC5
|
ATP binding cassette subfamily C member 5
|
|
chr19_-_36379185
Show fit
|
2.51 |
ENST00000270001.12
|
ZFP14
|
ZFP14 zinc finger protein
|
|
chr5_+_178941186
Show fit
|
2.51 |
ENST00000320129.7
ENST00000519564.2
|
ZNF454
|
zinc finger protein 454
|
|
chr16_-_10182754
Show fit
|
2.50 |
ENST00000396573.6
ENST00000675398.1
|
GRIN2A
|
glutamate ionotropic receptor NMDA type subunit 2A
|
|
chr3_+_156142962
Show fit
|
2.48 |
ENST00000471742.5
|
KCNAB1
|
potassium voltage-gated channel subfamily A member regulatory beta subunit 1
|
|
chrX_+_123859976
Show fit
|
2.41 |
ENST00000371199.8
|
XIAP
|
X-linked inhibitor of apoptosis
|
|
chr4_+_70195719
Show fit
|
2.36 |
ENST00000683306.1
|
ODAM
|
odontogenic, ameloblast associated
|
|
chr6_+_125749623
Show fit
|
2.34 |
ENST00000368364.4
|
HEY2
|
hes related family bHLH transcription factor with YRPW motif 2
|
|
chr10_+_122560679
Show fit
|
2.27 |
ENST00000657942.1
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr17_-_16215520
Show fit
|
2.21 |
ENST00000582357.5
ENST00000436828.5
ENST00000268712.8
ENST00000411510.5
ENST00000395857.7
|
NCOR1
|
nuclear receptor corepressor 1
|
|
chr10_+_122560639
Show fit
|
2.18 |
ENST00000344338.7
ENST00000330163.8
ENST00000652446.2
ENST00000666315.1
ENST00000368955.7
ENST00000368909.7
ENST00000368956.6
ENST00000619379.1
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr17_+_31094880
Show fit
|
2.18 |
ENST00000487476.5
ENST00000356175.7
|
NF1
|
neurofibromin 1
|
|
chr19_+_39997031
Show fit
|
2.11 |
ENST00000599504.5
ENST00000596894.5
ENST00000601138.5
ENST00000347077.9
ENST00000600094.5
|
ZNF546
|
zinc finger protein 546
|
|
chr15_-_33194696
Show fit
|
2.06 |
ENST00000320930.7
ENST00000616417.5
|
FMN1
|
formin 1
|
|
chr14_-_92748570
Show fit
|
2.05 |
ENST00000553918.1
ENST00000555699.5
ENST00000334869.9
ENST00000553802.5
ENST00000554397.5
ENST00000554919.5
ENST00000554080.5
ENST00000553371.1
ENST00000557434.5
ENST00000393218.6
|
LGMN
|
legumain
|
|
chr7_+_90709816
Show fit
|
2.04 |
ENST00000436577.3
|
CDK14
|
cyclin dependent kinase 14
|
|
chr3_+_169773384
Show fit
|
2.00 |
ENST00000349841.10
ENST00000356716.8
|
MYNN
|
myoneurin
|
|
chr7_+_94394886
Show fit
|
1.96 |
ENST00000297268.11
ENST00000620463.1
|
COL1A2
|
collagen type I alpha 2 chain
|
|
chr10_+_122560751
Show fit
|
1.86 |
ENST00000338354.10
ENST00000664692.1
ENST00000653442.1
ENST00000664974.1
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr7_+_90709530
Show fit
|
1.85 |
ENST00000406263.5
|
CDK14
|
cyclin dependent kinase 14
|
|
chr19_+_3094348
Show fit
|
1.82 |
ENST00000078429.9
|
GNA11
|
G protein subunit alpha 11
|
|
chr3_+_46407558
Show fit
|
1.81 |
ENST00000357392.4
ENST00000400880.3
ENST00000433848.1
|
CCRL2
|
C-C motif chemokine receptor like 2
|
|
chr1_+_43389889
Show fit
|
1.68 |
ENST00000562955.2
ENST00000634258.3
|
SZT2
|
SZT2 subunit of KICSTOR complex
|
|
chr9_+_99821846
Show fit
|
1.61 |
ENST00000338488.8
ENST00000618101.4
|
NR4A3
|
nuclear receptor subfamily 4 group A member 3
|
|
chr1_-_147172456
Show fit
|
1.61 |
ENST00000254101.4
|
PRKAB2
|
protein kinase AMP-activated non-catalytic subunit beta 2
|
|
chr2_+_48568981
Show fit
|
1.60 |
ENST00000394754.5
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough
|
|
chr5_+_141412979
Show fit
|
1.60 |
ENST00000612503.1
ENST00000398610.3
|
PCDHGA10
|
protocadherin gamma subfamily A, 10
|
|
chr2_+_3595049
Show fit
|
1.59 |
ENST00000236693.11
ENST00000349077.9
|
COLEC11
|
collectin subfamily member 11
|
|
chr9_-_95507416
Show fit
|
1.54 |
ENST00000429896.6
|
PTCH1
|
patched 1
|
|
chr7_+_90709231
Show fit
|
1.52 |
ENST00000446790.5
ENST00000265741.7
|
CDK14
|
cyclin dependent kinase 14
|
|
chr1_-_151008365
Show fit
|
1.46 |
ENST00000361936.9
ENST00000361738.11
|
MINDY1
|
MINDY lysine 48 deubiquitinase 1
|
|
chr2_-_79087986
Show fit
|
1.46 |
ENST00000305089.8
|
REG1B
|
regenerating family member 1 beta
|
|
chr3_+_171843337
Show fit
|
1.44 |
ENST00000334567.9
ENST00000619900.4
ENST00000450693.1
|
TMEM212
|
transmembrane protein 212
|
|
chr6_-_32224060
Show fit
|
1.40 |
ENST00000375023.3
|
NOTCH4
|
notch receptor 4
|
|
chr5_+_178895892
Show fit
|
1.39 |
ENST00000520660.5
ENST00000361362.7
ENST00000520805.5
|
ZFP2
|
ZFP2 zinc finger protein
|
|
chr12_+_57772587
Show fit
|
1.37 |
ENST00000300209.13
|
EEF1AKMT3
|
EEF1A lysine methyltransferase 3
|
|
chr18_+_13218195
Show fit
|
1.32 |
ENST00000679167.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4
|
|
chr9_-_14722725
Show fit
|
1.30 |
ENST00000380911.4
|
CER1
|
cerberus 1, DAN family BMP antagonist
|
|
chr17_+_31094969
Show fit
|
1.30 |
ENST00000431387.8
ENST00000358273.9
|
NF1
|
neurofibromin 1
|
|
chr3_-_45915698
Show fit
|
1.27 |
ENST00000539217.5
|
LZTFL1
|
leucine zipper transcription factor like 1
|
|
chr21_-_30166782
Show fit
|
1.27 |
ENST00000286808.5
|
CLDN17
|
claudin 17
|
|
chr14_-_52950992
Show fit
|
1.26 |
ENST00000343279.8
ENST00000399304.7
ENST00000395631.6
ENST00000341590.8
|
FERMT2
|
fermitin family member 2
|
|
chr16_+_53703963
Show fit
|
1.25 |
ENST00000636218.1
ENST00000637001.1
ENST00000471389.6
ENST00000637969.1
ENST00000640179.1
|
FTO
|
FTO alpha-ketoglutarate dependent dioxygenase
|
|
chr12_-_89656093
Show fit
|
1.23 |
ENST00000359142.7
|
ATP2B1
|
ATPase plasma membrane Ca2+ transporting 1
|
|
chr12_-_89656051
Show fit
|
1.23 |
ENST00000261173.6
|
ATP2B1
|
ATPase plasma membrane Ca2+ transporting 1
|
|
chr2_-_144516154
Show fit
|
1.19 |
ENST00000637304.1
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chrX_-_119605870
Show fit
|
1.16 |
ENST00000542113.3
|
NKRF
|
NFKB repressing factor
|
|
chr12_+_6200759
Show fit
|
1.16 |
ENST00000645565.1
ENST00000382515.7
|
CD9
|
CD9 molecule
|
|
chr8_+_106726012
Show fit
|
1.13 |
ENST00000449762.6
ENST00000297447.10
|
OXR1
|
oxidation resistance 1
|
|
chr9_+_706841
Show fit
|
1.13 |
ENST00000382293.7
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr16_-_70685791
Show fit
|
1.11 |
ENST00000616026.4
|
MTSS2
|
MTSS I-BAR domain containing 2
|
|
chr19_+_50025714
Show fit
|
1.10 |
ENST00000598809.5
ENST00000595661.5
ENST00000391821.6
|
ZNF473
|
zinc finger protein 473
|
|
chr20_+_31475278
Show fit
|
1.04 |
ENST00000201979.3
|
REM1
|
RRAD and GEM like GTPase 1
|
|
chrX_-_70260199
Show fit
|
1.03 |
ENST00000374519.4
|
P2RY4
|
pyrimidinergic receptor P2Y4
|
|
chr10_-_95441015
Show fit
|
1.01 |
ENST00000371241.5
ENST00000354106.7
ENST00000371239.5
ENST00000361941.7
ENST00000277982.9
ENST00000371245.7
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr19_+_38899946
Show fit
|
1.01 |
ENST00000572515.5
ENST00000313582.6
ENST00000575359.5
|
NFKBIB
|
NFKB inhibitor beta
|
|
chr16_-_53703883
Show fit
|
1.00 |
ENST00000262135.9
ENST00000564374.5
ENST00000566096.5
|
RPGRIP1L
|
RPGRIP1 like
|
|
chr22_+_39901075
Show fit
|
0.97 |
ENST00000344138.9
|
GRAP2
|
GRB2 related adaptor protein 2
|
|
chr2_-_144516397
Show fit
|
0.96 |
ENST00000638128.1
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr5_-_138178599
Show fit
|
0.94 |
ENST00000454473.5
ENST00000418329.5
ENST00000254900.10
ENST00000230901.9
ENST00000402931.5
ENST00000411594.6
ENST00000430331.1
|
BRD8
|
bromodomain containing 8
|
|
chr4_-_119628007
Show fit
|
0.94 |
ENST00000420633.1
ENST00000394439.5
|
PDE5A
|
phosphodiesterase 5A
|
|
chr3_+_194136138
Show fit
|
0.93 |
ENST00000232424.4
|
HES1
|
hes family bHLH transcription factor 1
|
|
chr17_-_41624541
Show fit
|
0.93 |
ENST00000540235.5
ENST00000311208.13
|
KRT17
|
keratin 17
|
|
chr5_-_88877967
Show fit
|
0.93 |
ENST00000508610.5
ENST00000636294.1
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chrX_-_49264668
Show fit
|
0.85 |
ENST00000455775.7
ENST00000652559.1
ENST00000376207.10
ENST00000557224.6
ENST00000684155.1
ENST00000376199.7
|
FOXP3
|
forkhead box P3
|
|
chr1_-_7940825
Show fit
|
0.84 |
ENST00000377507.8
|
TNFRSF9
|
TNF receptor superfamily member 9
|
|
chr1_-_92486049
Show fit
|
0.83 |
ENST00000427103.5
|
GFI1
|
growth factor independent 1 transcriptional repressor
|
|
chr16_-_53703810
Show fit
|
0.83 |
ENST00000569716.1
ENST00000562588.5
ENST00000621565.5
ENST00000562230.5
ENST00000563746.5
ENST00000568653.7
ENST00000647211.2
|
RPGRIP1L
|
RPGRIP1 like
|
|
chrX_+_19343893
Show fit
|
0.82 |
ENST00000540249.5
ENST00000545074.5
ENST00000379806.9
ENST00000423505.5
ENST00000422285.7
ENST00000417819.5
ENST00000355808.9
ENST00000379805.3
|
PDHA1
|
pyruvate dehydrogenase E1 subunit alpha 1
|
|
chr6_-_75493629
Show fit
|
0.80 |
ENST00000393004.6
|
FILIP1
|
filamin A interacting protein 1
|
|
chr11_+_118607579
Show fit
|
0.78 |
ENST00000530708.4
|
PHLDB1
|
pleckstrin homology like domain family B member 1
|
|
chr4_+_26319636
Show fit
|
0.77 |
ENST00000342295.6
ENST00000506956.5
ENST00000512671.6
ENST00000345843.8
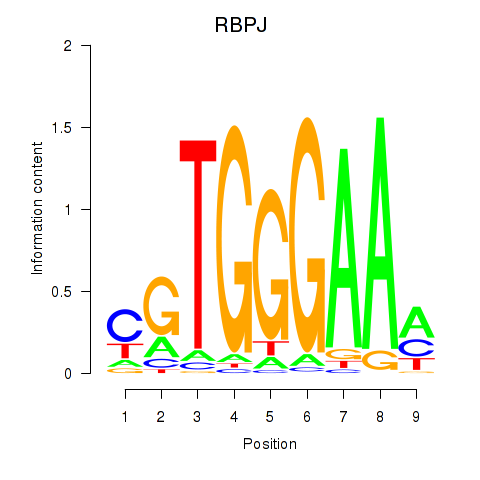
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region
|
|
chrX_+_147911910
Show fit
|
0.77 |
ENST00000370475.9
|
FMR1
|
FMRP translational regulator 1
|
|
chr16_+_23302292
Show fit
|
0.73 |
ENST00000343070.7
|
SCNN1B
|
sodium channel epithelial 1 subunit beta
|
|
chr11_-_8938211
Show fit
|
0.66 |
ENST00000531618.1
|
ASCL3
|
achaete-scute family bHLH transcription factor 3
|
|
chr3_-_87276577
Show fit
|
0.63 |
ENST00000344265.8
ENST00000350375.7
|
POU1F1
|
POU class 1 homeobox 1
|
|
chr1_-_39639626
Show fit
|
0.61 |
ENST00000372852.4
|
HEYL
|
hes related family bHLH transcription factor with YRPW motif like
|
|
chr15_+_40405787
Show fit
|
0.58 |
ENST00000610693.5
ENST00000479013.7
ENST00000487418.8
|
IVD
|
isovaleryl-CoA dehydrogenase
|
|
chr1_+_174799895
Show fit
|
0.56 |
ENST00000489615.5
|
RABGAP1L
|
RAB GTPase activating protein 1 like
|
|
chr3_+_69763549
Show fit
|
0.56 |
ENST00000472437.5
|
MITF
|
melanocyte inducing transcription factor
|
|
chrX_+_147911943
Show fit
|
0.55 |
ENST00000621453.4
ENST00000218200.12
ENST00000370471.7
ENST00000440235.6
ENST00000370477.5
ENST00000621987.4
|
FMR1
|
FMRP translational regulator 1
|
|
chr20_+_2652622
Show fit
|
0.54 |
ENST00000329276.10
ENST00000445139.1
|
NOP56
|
NOP56 ribonucleoprotein
|
|
chr11_-_124935973
Show fit
|
0.53 |
ENST00000298251.5
|
HEPACAM
|
hepatic and glial cell adhesion molecule
|
|
chr5_+_132673983
Show fit
|
0.52 |
ENST00000622422.1
ENST00000231449.7
ENST00000350025.2
|
IL4
|
interleukin 4
|
|
chr12_-_4649043
Show fit
|
0.50 |
ENST00000545990.6
ENST00000228850.6
|
AKAP3
|
A-kinase anchoring protein 3
|
|
chr7_-_16465728
Show fit
|
0.50 |
ENST00000307068.5
|
SOSTDC1
|
sclerostin domain containing 1
|
|
chr21_+_37420299
Show fit
|
0.50 |
ENST00000455097.6
ENST00000643854.1
ENST00000645424.1
ENST00000642309.1
ENST00000645774.1
ENST00000398956.2
|
DYRK1A
|
dual specificity tyrosine phosphorylation regulated kinase 1A
|
|
chr12_-_7695752
Show fit
|
0.48 |
ENST00000329913.4
|
GDF3
|
growth differentiation factor 3
|
|
chr11_-_64744993
Show fit
|
0.44 |
ENST00000377485.5
|
RASGRP2
|
RAS guanyl releasing protein 2
|
|
chr17_-_2336435
Show fit
|
0.44 |
ENST00000301364.10
ENST00000576112.2
|
TSR1
|
TSR1 ribosome maturation factor
|
|
chr7_+_76461676
Show fit
|
0.43 |
ENST00000425780.5
ENST00000456590.5
ENST00000451769.5
ENST00000324432.9
ENST00000457529.5
ENST00000446600.5
ENST00000430490.7
ENST00000413936.6
ENST00000423646.5
ENST00000438930.5
|
DTX2
|
deltex E3 ubiquitin ligase 2
|
|
chr7_-_27130182
Show fit
|
0.42 |
ENST00000511914.1
|
HOXA4
|
homeobox A4
|
|
chr14_-_21526391
Show fit
|
0.36 |
ENST00000611430.4
|
SALL2
|
spalt like transcription factor 2
|
|
chr12_-_12684490
Show fit
|
0.35 |
ENST00000540510.1
|
GPR19
|
G protein-coupled receptor 19
|
|
chr1_-_84893166
Show fit
|
0.27 |
ENST00000370611.4
|
LPAR3
|
lysophosphatidic acid receptor 3
|
|
chr2_-_144430934
Show fit
|
0.23 |
ENST00000638087.1
ENST00000638007.1
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr1_+_147902789
Show fit
|
0.23 |
ENST00000369235.2
|
GJA8
|
gap junction protein alpha 8
|
|
chr15_-_52529050
Show fit
|
0.21 |
ENST00000399231.7
|
MYO5A
|
myosin VA
|
|
chr6_+_106087580
Show fit
|
0.18 |
ENST00000424894.1
ENST00000648754.1
|
PRDM1
|
PR/SET domain 1
|
|
chr20_+_46008900
Show fit
|
0.18 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9
|
|
chr17_+_40318237
Show fit
|
0.13 |
ENST00000394089.6
ENST00000425707.7
|
RARA
|
retinoic acid receptor alpha
|
|
chr10_+_31319125
Show fit
|
0.11 |
ENST00000320985.14
ENST00000560721.6
ENST00000558440.5
ENST00000424869.6
ENST00000542815.7
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chrX_-_53686710
Show fit
|
0.06 |
ENST00000262854.11
|
HUWE1
|
HECT, UBA and WWE domain containing E3 ubiquitin protein ligase 1
|
|
chr7_-_95324524
Show fit
|
0.04 |
ENST00000222381.8
|
PON1
|
paraoxonase 1
|
|
chr1_-_43389768
Show fit
|
0.04 |
ENST00000372455.4
ENST00000372457.9
ENST00000290663.10
|
MED8
|
mediator complex subunit 8
|
|
chr11_-_83682385
Show fit
|
0.03 |
ENST00000426717.6
|
DLG2
|
discs large MAGUK scaffold protein 2
|
|
chr19_+_38899680
Show fit
|
0.02 |
ENST00000576510.5
ENST00000392079.7
|
NFKBIB
|
NFKB inhibitor beta
|
|
chr11_-_83682414
Show fit
|
0.01 |
ENST00000404783.7
|
DLG2
|
discs large MAGUK scaffold protein 2
|
|
chr8_-_100336184
Show fit
|
0.01 |
ENST00000519527.5
ENST00000522369.5
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase
|