Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
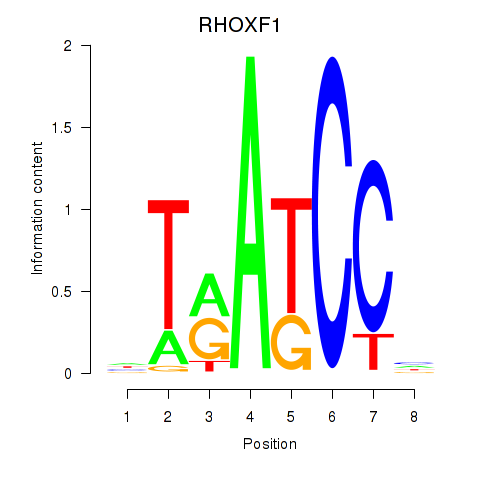
Results for RHOXF1
Z-value: 1.31
Transcription factors associated with RHOXF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RHOXF1
|
ENSG00000101883.5 | RHOXF1 |
Activity profile of RHOXF1 motif
Sorted Z-values of RHOXF1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RHOXF1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.8 | 32.4 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 10.2 | 30.5 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 9.8 | 39.1 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 6.9 | 20.8 | GO:0071140 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 6.3 | 31.6 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 6.3 | 25.2 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 4.9 | 63.5 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 4.3 | 13.0 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 4.0 | 12.0 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 3.9 | 3.9 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 3.8 | 15.3 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 3.7 | 26.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 3.4 | 16.9 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 2.6 | 7.8 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 2.4 | 7.2 | GO:0051941 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 2.3 | 6.9 | GO:0035759 | mesangial cell-matrix adhesion(GO:0035759) dendritic cell dendrite assembly(GO:0097026) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 2.2 | 11.0 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 2.1 | 8.6 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 2.1 | 10.5 | GO:0030070 | insulin processing(GO:0030070) |
| 2.1 | 10.4 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 2.0 | 8.2 | GO:0006562 | proline catabolic process(GO:0006562) |
| 2.0 | 6.1 | GO:0015847 | putrescine transport(GO:0015847) |
| 2.0 | 6.0 | GO:0070846 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 2.0 | 9.9 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 2.0 | 5.9 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 2.0 | 9.8 | GO:1902612 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 1.9 | 5.8 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 1.9 | 5.8 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 1.9 | 17.0 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 1.9 | 13.0 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 1.9 | 3.7 | GO:0002329 | pre-B cell differentiation(GO:0002329) pre-B cell allelic exclusion(GO:0002331) |
| 1.8 | 10.9 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 1.8 | 7.3 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 1.8 | 7.3 | GO:2000909 | regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 1.8 | 7.2 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 1.8 | 19.5 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 1.7 | 7.0 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 1.7 | 1.7 | GO:0035625 | epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 1.7 | 18.6 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 1.6 | 6.6 | GO:1900104 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 1.6 | 3.2 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 1.5 | 6.1 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 1.5 | 8.9 | GO:0018343 | protein farnesylation(GO:0018343) |
| 1.5 | 10.2 | GO:0048241 | epinephrine transport(GO:0048241) |
| 1.4 | 4.3 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 1.4 | 4.3 | GO:0071373 | cellular response to luteinizing hormone stimulus(GO:0071373) |
| 1.4 | 5.6 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 1.4 | 4.2 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 1.4 | 4.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) fourth ventricle development(GO:0021592) third ventricle development(GO:0021678) |
| 1.3 | 4.0 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 1.3 | 3.9 | GO:0018874 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 1.3 | 15.6 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 1.3 | 5.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 1.3 | 8.8 | GO:0034371 | positive regulation of sequestering of triglyceride(GO:0010890) chylomicron remodeling(GO:0034371) |
| 1.3 | 5.0 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 1.3 | 11.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 1.2 | 3.7 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) glomerular visceral epithelial cell migration(GO:0090521) |
| 1.2 | 9.9 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 1.2 | 3.7 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 1.2 | 7.3 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 1.2 | 3.6 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 1.2 | 18.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 1.2 | 3.6 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 1.2 | 4.8 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 1.2 | 8.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 1.2 | 4.7 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 1.2 | 5.9 | GO:0044856 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 1.2 | 4.7 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 1.2 | 4.7 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 1.1 | 3.4 | GO:1900081 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 1.1 | 19.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 1.1 | 4.5 | GO:1990834 | response to odorant(GO:1990834) |
| 1.1 | 2.2 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 1.1 | 5.6 | GO:0030035 | microspike assembly(GO:0030035) |
| 1.1 | 4.5 | GO:0009305 | protein biotinylation(GO:0009305) histone biotinylation(GO:0071110) |
| 1.1 | 3.3 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 1.1 | 3.2 | GO:0071529 | cementum mineralization(GO:0071529) |
| 1.1 | 4.3 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 1.1 | 3.2 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) |
| 1.1 | 7.4 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 1.1 | 3.2 | GO:1901874 | negative regulation of post-translational protein modification(GO:1901874) |
| 1.0 | 4.2 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 1.0 | 4.1 | GO:0090299 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 1.0 | 3.1 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 1.0 | 5.1 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 1.0 | 2.0 | GO:0014806 | smooth muscle hyperplasia(GO:0014806) |
| 1.0 | 4.0 | GO:1900191 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 1.0 | 3.0 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 1.0 | 5.0 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 1.0 | 7.9 | GO:0006477 | protein sulfation(GO:0006477) |
| 1.0 | 3.9 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 1.0 | 2.9 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 1.0 | 2.9 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 1.0 | 7.6 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 1.0 | 6.7 | GO:0035290 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.9 | 10.4 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.9 | 3.7 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.9 | 8.4 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.9 | 11.1 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.9 | 25.8 | GO:0050912 | detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.9 | 3.7 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.9 | 2.7 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.9 | 8.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.9 | 4.5 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.9 | 8.0 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.9 | 1.8 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.9 | 38.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.9 | 2.6 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.9 | 7.8 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.9 | 2.6 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.9 | 10.2 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.8 | 16.8 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.8 | 5.9 | GO:0097398 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.8 | 2.5 | GO:0048319 | mesodermal cell migration(GO:0008078) axial mesoderm morphogenesis(GO:0048319) |
| 0.8 | 4.2 | GO:0010816 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.8 | 3.3 | GO:1904980 | positive regulation of endosome organization(GO:1904980) |
| 0.8 | 23.1 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.8 | 4.9 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.8 | 1.6 | GO:0002537 | nitric oxide production involved in inflammatory response(GO:0002537) |
| 0.8 | 0.8 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.8 | 10.4 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.8 | 3.2 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.8 | 5.6 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.8 | 15.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.8 | 3.1 | GO:0033122 | negative regulation of cyclic nucleotide catabolic process(GO:0030806) negative regulation of cAMP catabolic process(GO:0030821) negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.8 | 3.9 | GO:0009804 | phenylpropanoid metabolic process(GO:0009698) coumarin metabolic process(GO:0009804) |
| 0.8 | 3.9 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.8 | 6.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.8 | 7.7 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.8 | 6.8 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.7 | 5.2 | GO:0045007 | depurination(GO:0045007) |
| 0.7 | 5.9 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 0.7 | 13.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.7 | 13.8 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.7 | 9.3 | GO:0046950 | cellular ketone body metabolic process(GO:0046950) |
| 0.7 | 2.9 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.7 | 7.1 | GO:1901538 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.7 | 2.9 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.7 | 10.6 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.7 | 2.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.7 | 7.0 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.7 | 2.8 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.7 | 4.8 | GO:0050666 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.7 | 3.5 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.7 | 2.1 | GO:0010840 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.7 | 2.1 | GO:0061183 | dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) positive regulation of dermatome development(GO:0061184) |
| 0.7 | 4.1 | GO:0061205 | alveolar primary septum development(GO:0061143) paramesonephric duct development(GO:0061205) |
| 0.7 | 6.8 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.7 | 4.8 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.7 | 11.5 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.7 | 5.3 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.7 | 4.0 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.6 | 7.1 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.6 | 8.8 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.6 | 13.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.6 | 6.8 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.6 | 11.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.6 | 4.3 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.6 | 1.2 | GO:2000722 | regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) |
| 0.6 | 2.4 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.6 | 1.8 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.6 | 4.8 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.6 | 5.9 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.6 | 44.3 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.6 | 2.9 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.6 | 3.5 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.6 | 12.7 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.6 | 3.5 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.6 | 3.4 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.6 | 3.4 | GO:0034380 | high-density lipoprotein particle assembly(GO:0034380) |
| 0.6 | 3.4 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.6 | 4.0 | GO:0098856 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.6 | 4.5 | GO:0015747 | urate transport(GO:0015747) |
| 0.6 | 1.7 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.5 | 1.6 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.5 | 8.7 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.5 | 3.7 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.5 | 2.1 | GO:1900158 | hormone-mediated apoptotic signaling pathway(GO:0008628) positive regulation of osteoclast proliferation(GO:0090290) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.5 | 2.1 | GO:0042640 | anagen(GO:0042640) |
| 0.5 | 9.0 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.5 | 2.6 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.5 | 1.6 | GO:0019878 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.5 | 4.7 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.5 | 2.0 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.5 | 7.6 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.5 | 3.0 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.5 | 2.5 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.5 | 9.9 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.5 | 3.4 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.5 | 2.4 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.5 | 2.9 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.5 | 6.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.5 | 4.3 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.5 | 2.4 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.5 | 3.8 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.5 | 2.8 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.5 | 0.9 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) regulation of protein lipidation(GO:1903059) positive regulation of protein lipidation(GO:1903061) |
| 0.5 | 2.3 | GO:0033504 | floor plate development(GO:0033504) |
| 0.5 | 2.3 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.5 | 3.7 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.5 | 4.5 | GO:0046959 | habituation(GO:0046959) |
| 0.5 | 1.4 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.4 | 0.9 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) negative regulation of gastrulation(GO:2000542) |
| 0.4 | 4.9 | GO:0098912 | membrane depolarization during AV node cell action potential(GO:0086045) membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.4 | 1.8 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) dorsal/ventral axon guidance(GO:0033563) |
| 0.4 | 4.0 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.4 | 0.9 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.4 | 5.7 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.4 | 3.4 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) endocardial cushion to mesenchymal transition(GO:0090500) |
| 0.4 | 43.5 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.4 | 2.6 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.4 | 3.8 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.4 | 3.4 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.4 | 2.9 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.4 | 2.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.4 | 1.7 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.4 | 2.9 | GO:0086100 | endothelin receptor signaling pathway(GO:0086100) |
| 0.4 | 4.5 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.4 | 7.0 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.4 | 4.9 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.4 | 10.2 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.4 | 2.0 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.4 | 4.4 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.4 | 12.4 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.4 | 1.6 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.4 | 19.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.4 | 5.4 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.4 | 0.8 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.4 | 5.8 | GO:1902221 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.4 | 8.7 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.4 | 3.0 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.4 | 8.9 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.4 | 2.2 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.4 | 21.5 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.4 | 2.9 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.4 | 7.2 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.4 | 3.9 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.4 | 7.4 | GO:0048070 | regulation of developmental pigmentation(GO:0048070) |
| 0.4 | 3.5 | GO:0033292 | T-tubule organization(GO:0033292) protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.4 | 1.1 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.3 | 2.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.3 | 1.7 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.3 | 3.4 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.3 | 4.8 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.3 | 2.7 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.3 | 3.4 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.3 | 5.7 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.3 | 2.3 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.3 | 1.7 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.3 | 1.7 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.3 | 2.3 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.3 | 5.8 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.3 | 8.0 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.3 | 4.8 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.3 | 1.0 | GO:0048865 | stem cell fate commitment(GO:0048865) |
| 0.3 | 5.7 | GO:0048240 | epithelial cilium movement(GO:0003351) sperm capacitation(GO:0048240) |
| 0.3 | 4.1 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.3 | 5.6 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.3 | 4.4 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.3 | 1.6 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.3 | 1.6 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.3 | 8.7 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.3 | 5.9 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.3 | 2.5 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.3 | 10.1 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.3 | 1.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.3 | 1.5 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.3 | 1.2 | GO:0032597 | B cell receptor transport within lipid bilayer(GO:0032595) B cell receptor transport into membrane raft(GO:0032597) protein transport out of membrane raft(GO:0032599) chemokine receptor transport out of membrane raft(GO:0032600) negative regulation of transforming growth factor beta3 production(GO:0032913) chemokine receptor transport within lipid bilayer(GO:0033606) |
| 0.3 | 3.3 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.3 | 1.5 | GO:0070778 | L-aspartate transport(GO:0070778) L-aspartate transmembrane transport(GO:0089712) |
| 0.3 | 11.1 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.3 | 1.5 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.3 | 0.9 | GO:1990918 | meiotic DNA double-strand break processing(GO:0000706) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.3 | 2.1 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.3 | 3.7 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.3 | 3.3 | GO:0045986 | negative regulation of smooth muscle contraction(GO:0045986) |
| 0.3 | 0.8 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.3 | 0.5 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.3 | 1.1 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.3 | 0.3 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.3 | 13.8 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.3 | 1.3 | GO:0021699 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.3 | 3.7 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.3 | 2.9 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.3 | 2.6 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.3 | 2.4 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.3 | 2.9 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.3 | 3.3 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.3 | 1.8 | GO:0002778 | antimicrobial peptide production(GO:0002775) antibacterial peptide production(GO:0002778) |
| 0.3 | 3.6 | GO:0051197 | positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.3 | 3.5 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.2 | 3.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.2 | 1.0 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.2 | 1.9 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.2 | 4.6 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.2 | 2.6 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.2 | 0.7 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of epithelial cell differentiation involved in kidney development(GO:2000698) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.2 | 1.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 2.3 | GO:0009584 | detection of visible light(GO:0009584) |
| 0.2 | 1.4 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.2 | 1.2 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.2 | 2.8 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.2 | 12.5 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.2 | 0.5 | GO:0061643 | chemorepulsion of dopaminergic neuron axon(GO:0036518) chemorepulsion of axon(GO:0061643) |
| 0.2 | 2.5 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.2 | 1.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 1.5 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.2 | 4.0 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 1.8 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.2 | 3.1 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.2 | 2.6 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.2 | 1.9 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.2 | 1.3 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.2 | 4.4 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.2 | 2.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 1.5 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.2 | 2.5 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.2 | 3.9 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.2 | 3.2 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.2 | 0.8 | GO:0002357 | defense response to tumor cell(GO:0002357) |
| 0.2 | 5.8 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.2 | 0.6 | GO:0035963 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 0.2 | 1.6 | GO:0034311 | sphingosine metabolic process(GO:0006670) diol metabolic process(GO:0034311) |
| 0.2 | 1.6 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.2 | 3.3 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.2 | 1.5 | GO:0007320 | insemination(GO:0007320) |
| 0.2 | 3.5 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.2 | 21.4 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.2 | 19.9 | GO:0070268 | cornification(GO:0070268) |
| 0.2 | 2.6 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.2 | 8.1 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.2 | 3.5 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.2 | 1.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.2 | 1.1 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.2 | 6.9 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.2 | 4.7 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.2 | 1.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 3.1 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.2 | 19.2 | GO:0005977 | glycogen metabolic process(GO:0005977) |
| 0.2 | 1.9 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.2 | 8.7 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.2 | 1.0 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.2 | 1.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 2.4 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.2 | 2.0 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.2 | 2.7 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.2 | 3.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.2 | 0.8 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.2 | 1.1 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.2 | 3.2 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.2 | 2.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.2 | 3.5 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.2 | 0.5 | GO:0060283 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.2 | 1.7 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.2 | 0.6 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.2 | 2.6 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 | 4.1 | GO:0043278 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.1 | 4.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 2.0 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 5.0 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 0.4 | GO:1990036 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 0.1 | 2.1 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.1 | 8.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 5.5 | GO:0030815 | negative regulation of cAMP metabolic process(GO:0030815) |
| 0.1 | 5.0 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.1 | 0.4 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 1.7 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 0.9 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.1 | 8.5 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 1.5 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 3.8 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 0.5 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.1 | 9.3 | GO:2000181 | negative regulation of angiogenesis(GO:0016525) negative regulation of blood vessel morphogenesis(GO:2000181) |
| 0.1 | 2.4 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.1 | 2.2 | GO:0071028 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.1 | 2.0 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.1 | 0.7 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.1 | 1.9 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.1 | 1.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.5 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 1.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 1.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 1.8 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.6 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.1 | 4.3 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 4.0 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.1 | 14.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 1.3 | GO:0048485 | sympathetic nervous system development(GO:0048485) |
| 0.1 | 1.7 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 1.3 | GO:0072678 | T cell migration(GO:0072678) |
| 0.1 | 2.0 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 0.3 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.7 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.1 | 1.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.4 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.1 | 0.2 | GO:0043380 | memory T cell differentiation(GO:0043379) regulation of memory T cell differentiation(GO:0043380) positive regulation of memory T cell differentiation(GO:0043382) |
| 0.1 | 6.1 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 3.9 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 0.5 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 1.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 2.1 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.1 | 6.3 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 1.5 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.1 | 1.5 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.1 | 0.3 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.1 | 3.0 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 0.3 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.1 | 0.7 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 0.8 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.1 | 1.3 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.1 | 0.5 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 1.2 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 | 0.9 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.1 | 1.0 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 0.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 1.4 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 1.0 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 1.8 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.4 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 0.7 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 2.2 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 2.0 | GO:0006835 | dicarboxylic acid transport(GO:0006835) |
| 0.0 | 0.3 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.1 | GO:1904844 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 2.2 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.2 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 1.3 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.8 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 2.1 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.1 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.7 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.7 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.8 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 0.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.2 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.4 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 1.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.8 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.2 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.8 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.5 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 1.1 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.5 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.5 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.3 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.1 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 | 0.2 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.8 | GO:0044243 | multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.6 | GO:0050830 | defense response to Gram-positive bacterium(GO:0050830) |
| 0.0 | 0.7 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.3 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.7 | GO:0032006 | regulation of TOR signaling(GO:0032006) |
| 0.0 | 0.6 | GO:0051591 | response to cAMP(GO:0051591) |
| 0.0 | 0.1 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.3 | 49.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 3.9 | 50.4 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 2.7 | 47.9 | GO:0042627 | chylomicron(GO:0042627) |
| 2.5 | 39.8 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 1.9 | 5.7 | GO:0097229 | sperm end piece(GO:0097229) |
| 1.9 | 28.3 | GO:0030478 | actin cap(GO:0030478) |
| 1.6 | 4.7 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 1.6 | 7.8 | GO:0044305 | calyx of Held(GO:0044305) |
| 1.4 | 6.8 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 1.3 | 14.0 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 1.3 | 8.9 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 1.2 | 30.4 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 1.0 | 9.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.9 | 6.6 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.9 | 12.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.9 | 11.3 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.8 | 2.5 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.8 | 4.2 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.8 | 11.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.8 | 4.6 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.8 | 2.3 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.8 | 3.0 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.7 | 5.2 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.7 | 7.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.7 | 13.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.7 | 12.9 | GO:0005922 | connexon complex(GO:0005922) |
| 0.7 | 10.6 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.7 | 4.8 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.7 | 3.3 | GO:0031673 | H zone(GO:0031673) |
| 0.7 | 3.9 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.6 | 13.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.6 | 2.9 | GO:0089701 | U2AF(GO:0089701) |
| 0.6 | 7.7 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.5 | 10.3 | GO:0036019 | endolysosome(GO:0036019) |
| 0.5 | 3.6 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.5 | 3.0 | GO:0034464 | BBSome(GO:0034464) |
| 0.5 | 2.5 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.5 | 22.8 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.5 | 8.9 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.5 | 6.8 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.5 | 4.3 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.5 | 3.4 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.5 | 3.2 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.5 | 5.5 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.4 | 3.7 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.4 | 8.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.4 | 4.8 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.4 | 27.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.4 | 2.3 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.4 | 1.5 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.4 | 39.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.4 | 3.7 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.4 | 4.9 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.3 | 4.5 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.3 | 2.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.3 | 4.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.3 | 1.6 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.3 | 2.9 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.3 | 4.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.3 | 7.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.3 | 1.9 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.3 | 3.8 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.3 | 1.5 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.3 | 11.0 | GO:0030673 | axolemma(GO:0030673) |
| 0.3 | 2.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.3 | 5.0 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.3 | 2.0 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.3 | 7.2 | GO:0043218 | compact myelin(GO:0043218) |
| 0.3 | 2.6 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.3 | 7.0 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.3 | 4.5 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.3 | 1.9 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.3 | 3.7 | GO:0044447 | axoneme part(GO:0044447) |
| 0.3 | 9.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.3 | 1.5 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.3 | 1.5 | GO:0000243 | commitment complex(GO:0000243) |
| 0.3 | 1.3 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.3 | 28.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.2 | 2.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.2 | 8.7 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 4.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 4.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 10.6 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.2 | 2.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 10.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 18.5 | GO:0043235 | receptor complex(GO:0043235) |
| 0.2 | 6.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.2 | 2.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 3.0 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.2 | 2.5 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.2 | 2.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 3.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 1.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.2 | 14.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 1.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.2 | 22.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 1.7 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 0.5 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.2 | 1.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.2 | 5.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 0.7 | GO:1990879 | CST complex(GO:1990879) |
| 0.2 | 7.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 1.8 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.2 | 16.9 | GO:0005814 | centriole(GO:0005814) |
| 0.2 | 3.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.2 | 5.2 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.2 | 1.4 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.2 | 0.5 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.2 | 3.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 0.9 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 24.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 4.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 4.3 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 42.8 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 8.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 2.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 1.6 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 0.5 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 11.7 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 2.6 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.1 | 0.5 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 1.6 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 0.9 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 16.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 204.2 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.1 | 136.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 2.4 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 10.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 1.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 0.5 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 0.5 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.4 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.1 | 0.3 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 4.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 1.9 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 1.4 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.1 | 4.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 9.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 8.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 4.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 4.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 1.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 2.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.3 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 2.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 4.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 1.1 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.4 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 1.5 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 0.9 | GO:0044439 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.5 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.2 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 33.1 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.6 | 19.8 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 5.1 | 46.3 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 5.0 | 34.8 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 4.4 | 13.2 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 4.4 | 26.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 4.2 | 12.5 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 4.0 | 12.0 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 3.7 | 14.6 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 3.3 | 9.9 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 3.2 | 13.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 3.2 | 13.0 | GO:0032810 | sterol response element binding(GO:0032810) |
| 3.0 | 20.8 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 2.9 | 8.8 | GO:0017129 | triglyceride binding(GO:0017129) |
| 2.7 | 8.2 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 2.6 | 10.2 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 2.5 | 7.6 | GO:0070052 | collagen V binding(GO:0070052) |
| 2.5 | 15.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 2.3 | 6.9 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 2.2 | 6.6 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 2.2 | 10.8 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 2.1 | 17.0 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 2.1 | 12.7 | GO:0051373 | FATZ binding(GO:0051373) |
| 2.1 | 8.5 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 2.1 | 10.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 2.0 | 6.1 | GO:0015203 | polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) |
| 2.0 | 15.7 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 1.9 | 7.8 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 1.9 | 1.9 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 1.8 | 7.2 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 1.7 | 10.4 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 1.7 | 5.2 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 1.7 | 5.2 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 1.7 | 5.1 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 1.7 | 6.7 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 1.6 | 6.2 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 1.6 | 4.7 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 1.6 | 18.7 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 1.5 | 6.1 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 1.5 | 10.2 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 1.4 | 2.9 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 1.4 | 7.2 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 1.4 | 9.9 | GO:0031433 | telethonin binding(GO:0031433) |
| 1.4 | 7.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 1.4 | 5.6 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 1.4 | 5.6 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 1.4 | 5.6 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 1.4 | 15.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 1.3 | 30.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 1.3 | 18.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 1.3 | 3.8 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 1.2 | 11.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 1.2 | 8.7 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 1.2 | 11.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 1.2 | 7.0 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 1.2 | 3.5 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 1.1 | 3.4 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 1.1 | 3.4 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 1.1 | 4.5 | GO:0004079 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 1.1 | 4.4 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 1.1 | 4.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 1.0 | 3.1 | GO:0016497 | substance K receptor activity(GO:0016497) |
| 1.0 | 6.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 1.0 | 3.9 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 1.0 | 3.9 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 1.0 | 15.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.9 | 5.7 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.9 | 3.7 | GO:0017089 | glycolipid transporter activity(GO:0017089) ceramide binding(GO:0097001) |
| 0.9 | 29.4 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.9 | 3.7 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.9 | 7.3 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.9 | 29.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.9 | 6.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.9 | 7.7 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.8 | 22.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.8 | 3.3 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.8 | 16.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.8 | 12.2 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.8 | 4.9 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.8 | 10.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.8 | 3.2 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.8 | 3.2 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.8 | 11.0 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.8 | 3.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.8 | 2.4 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.8 | 16.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.8 | 4.7 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.8 | 3.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.7 | 5.1 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.7 | 2.9 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.7 | 3.6 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.7 | 2.9 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.7 | 4.3 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.7 | 3.5 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.7 | 6.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.7 | 58.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.7 | 3.5 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.7 | 5.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.7 | 8.9 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.7 | 2.0 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.7 | 3.4 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.7 | 10.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.7 | 4.7 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.7 | 2.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.7 | 2.0 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.7 | 7.8 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.6 | 5.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.6 | 1.9 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.6 | 3.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.6 | 3.7 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.6 | 12.8 | GO:0031005 | filamin binding(GO:0031005) |
| 0.6 | 8.5 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.6 | 3.0 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.6 | 4.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.6 | 4.2 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.6 | 3.6 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.6 | 2.4 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.6 | 2.4 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.6 | 1.8 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.6 | 1.8 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.6 | 8.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.6 | 3.5 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.6 | 5.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.6 | 8.6 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.6 | 5.0 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.6 | 7.8 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.6 | 3.9 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.5 | 9.9 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.5 | 1.6 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.5 | 4.9 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.5 | 3.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.5 | 4.8 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.5 | 11.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.5 | 1.6 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.5 | 1.5 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.5 | 9.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.5 | 12.7 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.5 | 1.5 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.5 | 11.1 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.5 | 3.0 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.5 | 4.5 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.5 | 6.0 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.5 | 2.0 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.5 | 1.9 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.5 | 16.4 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.5 | 5.7 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.5 | 13.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.5 | 1.9 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.5 | 7.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.5 | 0.5 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.5 | 2.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.5 | 2.8 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.5 | 1.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.5 | 2.7 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.5 | 2.3 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.4 | 3.0 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.4 | 2.6 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.4 | 43.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.4 | 6.7 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.4 | 42.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.4 | 4.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.4 | 2.0 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.4 | 7.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.4 | 4.4 | GO:0004568 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.4 | 8.9 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.4 | 1.9 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.4 | 2.3 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.4 | 1.1 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.4 | 2.2 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.4 | 4.5 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.4 | 29.9 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.4 | 3.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.4 | 6.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.4 | 7.6 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.4 | 4.0 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.4 | 2.2 | GO:0008310 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.3 | 9.9 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.3 | 2.7 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.3 | 7.0 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.3 | 6.0 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.3 | 8.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.3 | 2.3 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.3 | 6.9 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.3 | 2.6 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.3 | 6.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.3 | 8.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.3 | 8.0 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.3 | 9.2 | GO:0008066 | glutamate receptor activity(GO:0008066) |
| 0.3 | 1.9 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.3 | 4.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.3 | 6.6 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.3 | 4.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.3 | 1.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.3 | 0.9 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.3 | 4.6 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.3 | 3.6 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.3 | 2.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.3 | 8.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.3 | 3.0 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.3 | 1.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.3 | 1.6 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.3 | 2.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.3 | 1.6 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.3 | 3.1 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.3 | 5.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.3 | 10.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.3 | 22.3 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.3 | 2.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.3 | 1.8 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.3 | 1.3 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.3 | 8.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 0.7 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 2.4 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.2 | 2.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 3.6 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.2 | 17.3 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.2 | 1.8 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 1.4 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.2 | 3.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.2 | 1.3 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.2 | 1.5 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.2 | 1.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.2 | 9.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.2 | 3.3 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.2 | 0.8 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.2 | 3.4 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.2 | 40.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.2 | 6.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 1.5 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.2 | 5.0 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 0.8 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) glycine:sodium symporter activity(GO:0015375) |
| 0.2 | 2.0 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.2 | 4.1 | GO:0051430 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 0.5 | GO:0005497 | androgen binding(GO:0005497) |
| 0.2 | 16.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.2 | 1.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.2 | 1.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 4.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.2 | 5.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.2 | 0.9 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.2 | 0.5 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.2 | 1.9 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.2 | 0.5 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.2 | 0.8 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.2 | 2.6 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 0.5 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.2 | 1.1 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 1.0 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.1 | 1.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 3.7 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 4.5 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 3.9 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 4.1 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.1 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 6.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 3.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 3.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.9 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 1.3 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 1.5 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 2.5 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 0.6 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 1.0 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 13.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 1.3 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.1 | 1.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 0.7 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 27.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.0 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 5.2 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 2.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 1.8 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.1 | 1.6 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 1.0 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 0.5 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.6 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 5.9 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 1.9 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.4 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.1 | 2.5 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 0.8 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.1 | 3.0 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 2.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.2 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 2.7 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 2.1 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 0.8 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.1 | 3.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 4.0 | GO:0020037 | heme binding(GO:0020037) |
| 0.1 | 1.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.7 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 2.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 3.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 2.5 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.5 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.1 | 8.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 3.0 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.1 | 1.4 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.1 | 1.2 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 6.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 7.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 1.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.9 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 2.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 3.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 0.6 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 1.7 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 0.8 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 2.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 3.7 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 3.5 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.5 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 2.9 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 2.7 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 1.1 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 1.5 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.7 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.9 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 2.8 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 3.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.8 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.9 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 1.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 2.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 2.8 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 1.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 2.6 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.3 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.2 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.4 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 1.0 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 84.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 1.0 | 10.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.5 | 10.8 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.5 | 6.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.5 | 129.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.4 | 8.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.4 | 5.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.4 | 9.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.4 | 6.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.4 | 5.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.3 | 15.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.3 | 5.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.3 | 3.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 14.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 3.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.2 | 50.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 7.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 5.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.2 | 3.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 12.4 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.2 | 8.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 13.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.2 | 3.7 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.2 | 12.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 1.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 2.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 4.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 6.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 2.8 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 4.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 2.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 7.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 4.6 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 2.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 2.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 4.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 1.5 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 3.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 1.6 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.1 | 4.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 1.3 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 7.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 35.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 1.0 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 2.6 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 2.9 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 2.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 0.4 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 0.7 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 0.8 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 3.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 1.0 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 2.9 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.1 | 1.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 2.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 0.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 0.5 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 9.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 2.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.5 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 1.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 2.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.1 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.5 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 65.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 1.7 | 17.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 1.5 | 37.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 1.2 | 3.5 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.9 | 13.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.9 | 22.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.8 | 17.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.7 | 15.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.7 | 12.9 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.7 | 4.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.7 | 11.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.6 | 6.8 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.6 | 11.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.6 | 11.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.6 | 12.6 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.6 | 9.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.6 | 12.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.5 | 6.0 | REACTOME OPSINS | Genes involved in Opsins |
| 0.5 | 17.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.5 | 3.0 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.5 | 22.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.5 | 27.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.5 | 43.3 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.5 | 19.8 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.5 | 3.2 | REACTOME GAB1 SIGNALOSOME | Genes involved in GAB1 signalosome |
| 0.5 | 10.5 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.4 | 10.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.4 | 8.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.4 | 4.6 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.4 | 5.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.4 | 7.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.4 | 0.8 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.4 | 28.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.4 | 6.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.4 | 5.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.4 | 13.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.4 | 16.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.4 | 51.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.3 | 4.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.3 | 9.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.3 | 10.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.3 | 4.6 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.3 | 16.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.3 | 5.9 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.3 | 3.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.3 | 15.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.3 | 0.8 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.3 | 9.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.3 | 17.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.3 | 2.9 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.3 | 4.9 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 12.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 2.3 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.2 | 4.5 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.2 | 6.0 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.2 | 4.8 | REACTOME IMMUNE SYSTEM | Genes involved in Immune System |
| 0.2 | 21.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 12.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 1.6 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.2 | 2.5 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.2 | 7.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 6.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 2.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 9.2 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.2 | 4.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 4.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.2 | 7.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.2 | 2.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 3.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 18.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 2.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 2.9 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.2 | 5.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 2.7 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.2 | 2.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.2 | 3.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.2 | 5.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 6.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 6.1 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 0.7 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.1 | 1.0 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 8.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 6.3 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.1 | 3.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 2.0 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 2.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 4.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.6 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 5.5 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 2.0 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 1.5 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 2.0 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 2.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 2.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.0 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.1 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 2.4 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 2.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 9.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 0.9 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 1.0 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 2.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.9 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 0.6 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 1.5 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 1.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 0.8 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 2.3 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 2.2 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.1 | 1.1 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.1 | 1.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 2.5 | REACTOME BIOLOGICAL OXIDATIONS | Genes involved in Biological oxidations |
| 0.1 | 4.9 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 3.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 0.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.9 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 7.0 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 1.4 | REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 7.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.2 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 12.5 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 2.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.0 | 0.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 1.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |