Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
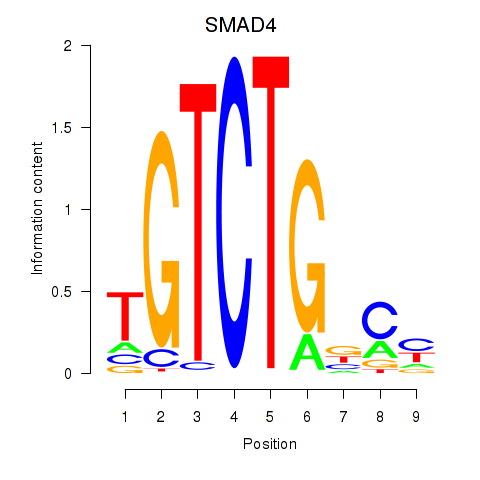
Results for SMAD4
Z-value: 1.92
Transcription factors associated with SMAD4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SMAD4
|
ENSG00000141646.14 | SMAD4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SMAD4 | hg38_v1_chr18_+_51030100_51030270 | 0.08 | 2.2e-01 | Click! |
Activity profile of SMAD4 motif
Sorted Z-values of SMAD4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SMAD4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.1 | 36.2 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 7.9 | 23.6 | GO:0071464 | cellular response to hydrostatic pressure(GO:0071464) |
| 7.7 | 7.7 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 6.3 | 18.8 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 6.2 | 24.9 | GO:0071231 | cellular response to folic acid(GO:0071231) |
| 6.2 | 18.5 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 5.6 | 33.5 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 4.9 | 19.6 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 4.9 | 14.7 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 4.8 | 81.7 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 4.8 | 14.3 | GO:0014873 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 4.7 | 14.1 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 4.4 | 31.0 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 4.4 | 13.2 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 4.3 | 13.0 | GO:0001300 | chronological cell aging(GO:0001300) |
| 4.3 | 13.0 | GO:0035793 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 4.3 | 12.9 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 4.2 | 4.2 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 4.2 | 12.6 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) mast cell proliferation(GO:0070662) |
| 3.9 | 27.4 | GO:0002774 | Fc receptor mediated inhibitory signaling pathway(GO:0002774) |
| 3.7 | 11.1 | GO:0035915 | pore formation in membrane of other organism(GO:0035915) |
| 3.6 | 10.7 | GO:2000547 | dendritic cell dendrite assembly(GO:0097026) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 3.4 | 10.3 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 3.4 | 20.4 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 3.3 | 10.0 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) |
| 3.2 | 25.6 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 3.0 | 9.1 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 2.9 | 14.7 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 2.9 | 14.6 | GO:0044856 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 2.9 | 8.7 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 2.9 | 20.0 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 2.8 | 11.0 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 2.8 | 13.8 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 2.7 | 10.9 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 2.6 | 7.9 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 2.6 | 2.6 | GO:2000137 | negative regulation of cell proliferation involved in heart morphogenesis(GO:2000137) |
| 2.5 | 7.6 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 2.4 | 9.8 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 2.4 | 36.4 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 2.4 | 7.3 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 2.3 | 7.0 | GO:0006311 | meiotic gene conversion(GO:0006311) |
| 2.3 | 9.3 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 2.3 | 6.9 | GO:0014813 | skeletal muscle satellite cell commitment(GO:0014813) |
| 2.2 | 6.7 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) beta-alanine metabolic process(GO:0019482) thymine metabolic process(GO:0019859) |
| 2.2 | 165.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 2.2 | 11.0 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 2.2 | 6.5 | GO:0060005 | vestibular reflex(GO:0060005) |
| 2.2 | 28.1 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 2.1 | 8.5 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 2.1 | 6.3 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 2.1 | 8.3 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 1.9 | 5.7 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 1.9 | 13.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 1.9 | 5.6 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 1.8 | 22.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 1.8 | 10.8 | GO:0008218 | bioluminescence(GO:0008218) |
| 1.8 | 5.3 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 1.8 | 5.3 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) positive regulation of somatostatin secretion(GO:0090274) |
| 1.8 | 3.5 | GO:0090264 | immune complex clearance(GO:0002434) immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) |
| 1.8 | 8.8 | GO:1904304 | regulation of gastro-intestinal system smooth muscle contraction(GO:1904304) positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 1.8 | 5.3 | GO:0014016 | noradrenergic neuron development(GO:0003358) neuroblast differentiation(GO:0014016) spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) regulation of timing of neuron differentiation(GO:0060164) olfactory pit development(GO:0060166) |
| 1.7 | 8.7 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 1.7 | 5.1 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 1.7 | 5.0 | GO:0019075 | virus maturation(GO:0019075) |
| 1.7 | 28.2 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 1.7 | 10.0 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 1.6 | 16.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 1.6 | 4.9 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 1.6 | 4.9 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 1.6 | 14.5 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 1.6 | 6.4 | GO:1902023 | L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 1.6 | 8.0 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 1.6 | 8.0 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 1.6 | 6.3 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 1.5 | 10.8 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 1.5 | 7.7 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 1.5 | 18.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 1.5 | 4.5 | GO:0098759 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 1.5 | 4.5 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 1.5 | 7.4 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 1.5 | 4.4 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 1.5 | 18.9 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 1.4 | 18.7 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 1.4 | 7.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 1.4 | 8.5 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 1.4 | 12.5 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 1.4 | 2.8 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 1.4 | 8.3 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 1.4 | 5.5 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) |
| 1.4 | 4.1 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 1.4 | 23.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 1.4 | 5.4 | GO:0009822 | alkaloid catabolic process(GO:0009822) calcitriol biosynthetic process from calciol(GO:0036378) |
| 1.3 | 12.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 1.3 | 2.6 | GO:0002884 | negative regulation of hypersensitivity(GO:0002884) |
| 1.3 | 29.9 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 1.3 | 5.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 1.3 | 20.1 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 1.3 | 8.8 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 1.3 | 1.3 | GO:0034756 | regulation of iron ion transport(GO:0034756) regulation of iron ion transmembrane transport(GO:0034759) response to iron ion starvation(GO:1990641) |
| 1.2 | 7.5 | GO:0006050 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 1.2 | 8.3 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 1.2 | 2.4 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 1.2 | 11.7 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 1.2 | 3.5 | GO:0061011 | hepatic duct development(GO:0061011) |
| 1.1 | 6.9 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 1.1 | 107.8 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 1.1 | 3.4 | GO:0060374 | mast cell differentiation(GO:0060374) |
| 1.1 | 10.0 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 1.1 | 119.6 | GO:0031295 | T cell costimulation(GO:0031295) |
| 1.1 | 3.2 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 1.1 | 3.2 | GO:0071529 | cementum mineralization(GO:0071529) |
| 1.1 | 3.2 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 1.1 | 12.7 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 1.1 | 3.2 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 1.1 | 2.1 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 1.0 | 3.1 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 1.0 | 4.1 | GO:2001034 | histone H3-K36 dimethylation(GO:0097676) positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 1.0 | 6.0 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 1.0 | 3.0 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 1.0 | 15.8 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 1.0 | 16.7 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 1.0 | 10.7 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 1.0 | 15.5 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.9 | 7.6 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.9 | 4.7 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.9 | 3.8 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.9 | 2.8 | GO:0051365 | leading edge cell differentiation(GO:0035026) cellular response to potassium ion starvation(GO:0051365) |
| 0.9 | 8.3 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.9 | 14.8 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.9 | 3.6 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
| 0.9 | 4.5 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.9 | 6.3 | GO:1901509 | branch elongation involved in ureteric bud branching(GO:0060681) regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.9 | 3.6 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.9 | 2.7 | GO:1990654 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.9 | 1.8 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.9 | 6.9 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 0.9 | 9.5 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.9 | 2.6 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.8 | 2.5 | GO:0072573 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) |
| 0.8 | 1.7 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.8 | 9.1 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.8 | 4.1 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.8 | 5.8 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.8 | 2.5 | GO:0035048 | splicing factor protein import into nucleus(GO:0035048) |
| 0.8 | 1.6 | GO:0021834 | embryonic olfactory bulb interneuron precursor migration(GO:0021831) chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 0.8 | 9.4 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.8 | 1.6 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.8 | 1.5 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.8 | 3.0 | GO:0009137 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.8 | 3.0 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.7 | 3.7 | GO:1903904 | negative regulation of establishment of T cell polarity(GO:1903904) negative regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001107) |
| 0.7 | 3.0 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.7 | 10.4 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.7 | 2.9 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.7 | 5.8 | GO:2000399 | negative regulation of T cell differentiation in thymus(GO:0033085) negative regulation of thymocyte aggregation(GO:2000399) |
| 0.7 | 4.3 | GO:0075044 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.7 | 5.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.7 | 2.9 | GO:0051582 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.7 | 24.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.7 | 2.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.7 | 4.2 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.7 | 3.5 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.7 | 5.6 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.7 | 6.3 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.7 | 10.3 | GO:0007530 | sex determination(GO:0007530) |
| 0.7 | 2.7 | GO:0051971 | positive regulation of transmission of nerve impulse(GO:0051971) |
| 0.7 | 3.4 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.7 | 4.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.7 | 5.4 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.7 | 3.4 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.7 | 3.3 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.7 | 3.9 | GO:2001206 | positive regulation of bone development(GO:1903012) positive regulation of osteoclast development(GO:2001206) |
| 0.7 | 3.9 | GO:0050893 | sensory processing(GO:0050893) |
| 0.6 | 8.4 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.6 | 4.5 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.6 | 4.5 | GO:0035093 | piRNA metabolic process(GO:0034587) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.6 | 3.9 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.6 | 17.2 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.6 | 8.9 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.6 | 4.5 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.6 | 11.3 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.6 | 1.9 | GO:2000566 | positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.6 | 1.9 | GO:0038156 | interleukin-5-mediated signaling pathway(GO:0038043) interleukin-3-mediated signaling pathway(GO:0038156) |
| 0.6 | 1.9 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.6 | 8.6 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.6 | 1.8 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.6 | 6.7 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.6 | 3.0 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.6 | 3.0 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.6 | 1.8 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.6 | 2.4 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.6 | 7.1 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.6 | 1.2 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.6 | 88.2 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.6 | 1.2 | GO:0070428 | nitric oxide production involved in inflammatory response(GO:0002537) negative regulation of interleukin-23 production(GO:0032707) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.6 | 6.4 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.6 | 15.7 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.6 | 4.0 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.6 | 4.0 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.6 | 1.7 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.6 | 4.5 | GO:0015014 | protein sulfation(GO:0006477) heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.6 | 2.2 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.6 | 2.8 | GO:0021699 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.5 | 4.9 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.5 | 2.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.5 | 5.8 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.5 | 2.7 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.5 | 2.6 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.5 | 3.7 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.5 | 6.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.5 | 2.6 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.5 | 4.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.5 | 3.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.5 | 13.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.5 | 2.0 | GO:0002268 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.5 | 4.6 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.5 | 3.0 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.5 | 7.0 | GO:1903798 | regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 0.5 | 10.1 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.5 | 11.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.5 | 1.0 | GO:0046643 | regulation of gamma-delta T cell activation(GO:0046643) positive regulation of gamma-delta T cell activation(GO:0046645) |
| 0.5 | 9.7 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.5 | 4.8 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.5 | 1.4 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.5 | 3.3 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.5 | 8.0 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.5 | 3.3 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.5 | 4.2 | GO:0035542 | endosomal vesicle fusion(GO:0034058) regulation of SNARE complex assembly(GO:0035542) positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.5 | 2.8 | GO:2000252 | negative regulation of feeding behavior(GO:2000252) |
| 0.5 | 1.8 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.5 | 9.1 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.5 | 2.3 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.5 | 1.8 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.5 | 6.3 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.5 | 2.7 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.5 | 2.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.5 | 4.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.4 | 27.3 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.4 | 1.3 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.4 | 8.9 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.4 | 4.0 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.4 | 3.5 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.4 | 12.7 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.4 | 3.0 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.4 | 10.8 | GO:0051412 | response to corticosterone(GO:0051412) |
| 0.4 | 1.3 | GO:0061743 | motor learning(GO:0061743) |
| 0.4 | 1.7 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.4 | 3.8 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.4 | 9.7 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.4 | 5.0 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.4 | 5.9 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.4 | 4.6 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.4 | 1.3 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.4 | 7.1 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.4 | 16.7 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.4 | 12.1 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.4 | 2.1 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.4 | 2.4 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.4 | 2.4 | GO:0061143 | alveolar primary septum development(GO:0061143) paramesonephric duct development(GO:0061205) |
| 0.4 | 7.9 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.4 | 4.0 | GO:0072540 | T-helper cell lineage commitment(GO:0002295) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) T-helper 17 cell lineage commitment(GO:0072540) |
| 0.4 | 9.4 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.4 | 13.3 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.4 | 9.7 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.4 | 3.5 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.4 | 7.4 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.4 | 25.5 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.4 | 6.1 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.4 | 6.8 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.4 | 29.4 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.4 | 3.0 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.4 | 2.9 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.4 | 1.1 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.4 | 2.9 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.4 | 1.4 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.4 | 3.5 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.3 | 1.7 | GO:0097155 | embryonic heart tube left/right pattern formation(GO:0060971) fasciculation of sensory neuron axon(GO:0097155) |
| 0.3 | 2.4 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.3 | 1.4 | GO:0032700 | negative regulation of interleukin-17 production(GO:0032700) |
| 0.3 | 5.7 | GO:0090312 | positive regulation of protein deacetylation(GO:0090312) |
| 0.3 | 2.4 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.3 | 3.7 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.3 | 4.3 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.3 | 3.0 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.3 | 1.9 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.3 | 1.3 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.3 | 3.8 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.3 | 15.6 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.3 | 6.2 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.3 | 2.5 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.3 | 1.8 | GO:0051945 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) regulation of peroxidase activity(GO:2000468) positive regulation of peroxidase activity(GO:2000470) |
| 0.3 | 3.9 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.3 | 3.9 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.3 | 3.8 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.3 | 3.2 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.3 | 4.0 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.3 | 2.6 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.3 | 12.8 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.3 | 1.7 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.3 | 1.7 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 0.3 | 2.2 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.3 | 3.0 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.3 | 5.0 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.3 | 5.2 | GO:0003170 | heart valve development(GO:0003170) |
| 0.3 | 3.0 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.3 | 1.4 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.3 | 3.5 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.3 | 4.0 | GO:0045821 | positive regulation of glycolytic process(GO:0045821) |
| 0.3 | 1.3 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.3 | 30.7 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.3 | 2.6 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.3 | 2.9 | GO:0010572 | positive regulation of platelet activation(GO:0010572) regulation of integrin activation(GO:0033623) |
| 0.3 | 6.2 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.3 | 5.4 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.3 | 1.3 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.3 | 1.0 | GO:1901098 | regulation of autophagosome maturation(GO:1901096) positive regulation of autophagosome maturation(GO:1901098) |
| 0.2 | 4.7 | GO:0002029 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) negative adaptation of signaling pathway(GO:0022401) |
| 0.2 | 7.4 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.2 | 7.3 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.2 | 0.4 | GO:0009820 | alkaloid metabolic process(GO:0009820) |
| 0.2 | 5.9 | GO:0034776 | response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 0.2 | 2.2 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.2 | 4.8 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.2 | 2.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 0.9 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.2 | 3.8 | GO:0050812 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.2 | 1.5 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.2 | 1.5 | GO:0090656 | t-circle formation(GO:0090656) |
| 0.2 | 5.0 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.2 | 2.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.2 | 2.7 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.2 | 0.8 | GO:0045408 | regulation of interleukin-6 biosynthetic process(GO:0045408) |
| 0.2 | 0.6 | GO:1901874 | negative regulation of post-translational protein modification(GO:1901874) |
| 0.2 | 3.1 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.2 | 4.6 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.2 | 5.0 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.2 | 0.8 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.2 | 7.2 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.2 | 1.7 | GO:0007320 | insemination(GO:0007320) |
| 0.2 | 7.0 | GO:0016233 | telomere capping(GO:0016233) |
| 0.2 | 5.8 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.2 | 9.1 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.2 | 2.9 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.2 | 0.9 | GO:0046641 | positive regulation of alpha-beta T cell proliferation(GO:0046641) |
| 0.2 | 5.0 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.2 | 3.3 | GO:0048536 | spleen development(GO:0048536) |
| 0.2 | 1.0 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.2 | 2.0 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.2 | 9.4 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.2 | 10.7 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.2 | 5.1 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.2 | 0.8 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 6.5 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.2 | 4.8 | GO:0015813 | acidic amino acid transport(GO:0015800) L-glutamate transport(GO:0015813) |
| 0.2 | 2.8 | GO:0006525 | arginine metabolic process(GO:0006525) |
| 0.2 | 3.1 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.2 | 5.7 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.2 | 0.6 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.2 | 1.5 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.2 | 2.1 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 3.0 | GO:0001964 | startle response(GO:0001964) |
| 0.1 | 6.7 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.1 | 0.7 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 5.2 | GO:0045453 | bone resorption(GO:0045453) |
| 0.1 | 1.8 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 2.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 4.4 | GO:0031640 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.1 | 3.1 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.1 | 6.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 1.9 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 0.4 | GO:2000302 | positive regulation of synaptic vesicle priming(GO:0010808) positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.1 | 1.6 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.1 | 2.2 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 2.7 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 1.4 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.1 | 8.7 | GO:0051899 | membrane depolarization(GO:0051899) |
| 0.1 | 3.1 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 1.2 | GO:0086091 | regulation of heart rate by cardiac conduction(GO:0086091) |
| 0.1 | 6.5 | GO:0042303 | molting cycle(GO:0042303) hair cycle(GO:0042633) |
| 0.1 | 6.0 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.1 | 3.4 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 0.2 | GO:0006029 | proteoglycan metabolic process(GO:0006029) |
| 0.1 | 0.7 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 5.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.4 | GO:0050746 | regulation of lipoprotein metabolic process(GO:0050746) |
| 0.1 | 3.1 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.1 | 6.2 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 1.8 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.1 | 1.5 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 4.6 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 5.0 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.1 | 0.7 | GO:0006568 | tryptophan metabolic process(GO:0006568) |
| 0.1 | 0.6 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 | 2.3 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.1 | 2.0 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 3.5 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.1 | 3.2 | GO:0032945 | negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) negative regulation of leukocyte proliferation(GO:0070664) |
| 0.1 | 3.0 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.1 | 4.5 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 0.3 | GO:0002829 | negative regulation of type 2 immune response(GO:0002829) negative regulation of interleukin-13 production(GO:0032696) |
| 0.1 | 4.2 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 0.6 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.1 | 0.8 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.1 | 1.3 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 6.6 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 2.1 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 2.1 | GO:0055010 | ventricular cardiac muscle tissue morphogenesis(GO:0055010) |
| 0.1 | 1.1 | GO:0048485 | sympathetic nervous system development(GO:0048485) |
| 0.1 | 2.6 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.1 | 2.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 1.4 | GO:0010644 | cell communication by electrical coupling(GO:0010644) |
| 0.1 | 2.3 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.1 | 0.8 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.1 | 0.9 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.1 | 0.3 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 3.8 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 2.9 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 1.8 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.1 | 0.7 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 7.9 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 2.0 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 3.1 | GO:0001523 | retinoid metabolic process(GO:0001523) |
| 0.1 | 4.2 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.1 | 0.3 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 5.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 0.3 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 | 0.9 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.1 | 2.5 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 0.7 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 1.5 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 2.6 | GO:0006835 | dicarboxylic acid transport(GO:0006835) |
| 0.0 | 2.8 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 2.8 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 1.4 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.1 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.0 | 0.3 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 | 2.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 20.5 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.5 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.6 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 3.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.7 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.8 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.4 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 0.4 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.5 | 123.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 5.1 | 81.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 4.9 | 14.7 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 4.6 | 27.8 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 4.3 | 13.0 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 3.7 | 29.9 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 3.0 | 161.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 2.6 | 10.2 | GO:0071942 | XPC complex(GO:0071942) |
| 2.5 | 12.5 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 2.4 | 14.5 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 2.0 | 6.0 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 1.9 | 5.7 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 1.8 | 19.6 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 1.8 | 7.0 | GO:0000801 | central element(GO:0000801) |
| 1.7 | 7.0 | GO:1990879 | CST complex(GO:1990879) |
| 1.7 | 15.6 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 1.7 | 22.1 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 1.7 | 5.0 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 1.6 | 14.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 1.6 | 20.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 1.5 | 8.8 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 1.4 | 10.0 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 1.4 | 9.7 | GO:0032010 | phagolysosome(GO:0032010) |
| 1.3 | 4.0 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 1.3 | 4.0 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 1.2 | 6.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 1.2 | 11.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 1.2 | 3.5 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 1.1 | 3.2 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 1.1 | 10.7 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 1.0 | 15.7 | GO:0030478 | actin cap(GO:0030478) |
| 1.0 | 5.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 1.0 | 24.9 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 1.0 | 11.4 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.9 | 1.8 | GO:0042599 | lamellar body(GO:0042599) |
| 0.9 | 17.1 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.8 | 5.1 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.8 | 20.0 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.8 | 4.7 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.8 | 3.1 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.8 | 14.7 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.7 | 13.7 | GO:0036038 | MKS complex(GO:0036038) |
| 0.7 | 1.4 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.7 | 10.6 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.7 | 16.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.6 | 3.8 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.6 | 6.7 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.6 | 3.6 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.6 | 4.8 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.6 | 2.4 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.6 | 3.5 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.6 | 41.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.6 | 2.3 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.6 | 9.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.6 | 5.0 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.5 | 2.0 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.5 | 3.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.5 | 4.5 | GO:0033391 | mRNA cap binding complex(GO:0005845) chromatoid body(GO:0033391) |
| 0.5 | 4.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.5 | 17.0 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.5 | 30.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.5 | 6.3 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.5 | 17.8 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.5 | 5.2 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.5 | 4.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.4 | 9.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.4 | 5.7 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.4 | 64.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.4 | 2.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.4 | 54.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.4 | 10.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.4 | 11.8 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.4 | 7.9 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.4 | 3.0 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.4 | 4.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.3 | 4.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.3 | 13.1 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.3 | 9.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.3 | 3.7 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.3 | 3.0 | GO:0045180 | basal cortex(GO:0045180) |
| 0.3 | 3.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 4.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.3 | 6.0 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.3 | 4.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.3 | 2.8 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.3 | 35.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.3 | 1.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.3 | 6.1 | GO:0005605 | basal lamina(GO:0005605) |
| 0.3 | 9.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.3 | 3.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.3 | 5.8 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.3 | 3.0 | GO:0070449 | elongin complex(GO:0070449) |
| 0.3 | 2.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.3 | 6.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.3 | 53.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.3 | 16.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.3 | 8.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.3 | 1.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.3 | 14.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.2 | 2.9 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.2 | 14.0 | GO:0043235 | receptor complex(GO:0043235) |
| 0.2 | 16.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.2 | 15.2 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.2 | 9.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 6.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 30.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.2 | 14.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 2.9 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.2 | 7.0 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.2 | 0.8 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 3.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.2 | 9.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.2 | 7.0 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.2 | 1.5 | GO:0016011 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.2 | 15.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.2 | 6.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 3.9 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 1.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.2 | 1.6 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.2 | 0.8 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 4.7 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 0.4 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 1.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 1.0 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 4.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 15.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 18.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 0.7 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 9.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.3 | GO:0033643 | host cell part(GO:0033643) |
| 0.1 | 19.6 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 2.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 2.0 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 2.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 3.2 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.1 | 10.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 0.6 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 8.2 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.1 | 6.6 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.1 | 2.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 19.4 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 2.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.8 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.9 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.9 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 5.2 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 3.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 0.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 9.0 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.1 | 2.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 5.2 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 1.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 0.7 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 4.1 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 7.8 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.1 | 61.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 3.7 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.1 | 0.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 2.4 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 5.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 9.0 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 14.8 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 24.9 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 5.6 | 33.5 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 5.1 | 20.5 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 4.9 | 19.7 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 4.9 | 14.7 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 4.6 | 54.8 | GO:0008430 | selenium binding(GO:0008430) |
| 4.4 | 61.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 4.1 | 33.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 3.4 | 27.4 | GO:0032396 | inhibitory MHC class I receptor activity(GO:0032396) |
| 3.3 | 13.2 | GO:0032810 | sterol response element binding(GO:0032810) |
| 3.1 | 9.4 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 3.1 | 9.2 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 2.9 | 8.8 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 2.8 | 14.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 2.8 | 11.0 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 2.7 | 81.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 2.7 | 10.9 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 2.6 | 10.3 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 2.6 | 154.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 2.6 | 20.4 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 2.5 | 7.6 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 2.2 | 15.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 2.2 | 6.6 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 2.2 | 28.1 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 2.0 | 23.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 1.9 | 39.4 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 1.8 | 9.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 1.8 | 57.9 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 1.8 | 5.4 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 1.8 | 7.2 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 1.8 | 10.7 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 1.8 | 31.9 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 1.7 | 3.5 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 1.7 | 10.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 1.7 | 5.1 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 1.7 | 10.2 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) |
| 1.7 | 15.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 1.7 | 5.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 1.7 | 8.3 | GO:0050436 | microfibril binding(GO:0050436) |
| 1.6 | 13.0 | GO:0005534 | galactose binding(GO:0005534) |
| 1.6 | 4.8 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 1.6 | 7.8 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 1.5 | 7.6 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 1.5 | 6.0 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 1.5 | 4.5 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 1.5 | 7.4 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 1.4 | 9.8 | GO:0030911 | TPR domain binding(GO:0030911) |
| 1.4 | 9.7 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 1.3 | 14.8 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 1.3 | 5.3 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 1.3 | 6.6 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 1.3 | 3.9 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 1.3 | 6.5 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 1.3 | 31.7 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 1.3 | 2.5 | GO:0001031 | RNA polymerase III type 1 promoter DNA binding(GO:0001030) RNA polymerase III type 2 promoter DNA binding(GO:0001031) 5S rDNA binding(GO:0080084) |
| 1.3 | 7.6 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 1.2 | 6.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 1.2 | 4.9 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 1.2 | 7.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 1.2 | 8.3 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 1.2 | 8.3 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 1.2 | 3.5 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 1.2 | 15.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 1.1 | 4.5 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 1.1 | 5.6 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 1.1 | 3.3 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 1.1 | 158.9 | GO:0003823 | antigen binding(GO:0003823) |
| 1.1 | 7.5 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 1.1 | 2.1 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 1.0 | 14.6 | GO:0045159 | myosin II binding(GO:0045159) |
| 1.0 | 4.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 1.0 | 4.0 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 1.0 | 14.0 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 1.0 | 21.0 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 1.0 | 3.0 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 1.0 | 5.0 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 1.0 | 2.9 | GO:0042806 | fucose binding(GO:0042806) |
| 1.0 | 5.7 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.9 | 4.7 | GO:0004368 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.9 | 3.8 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.9 | 2.8 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.9 | 7.4 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.9 | 12.7 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.9 | 12.7 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.9 | 2.7 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.9 | 6.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.9 | 4.4 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.9 | 5.1 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.8 | 5.0 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.8 | 1.7 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.8 | 3.2 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.8 | 2.4 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.8 | 4.8 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.8 | 2.4 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.8 | 1.6 | GO:0019841 | retinol binding(GO:0019841) |
| 0.8 | 7.7 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.8 | 5.3 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.8 | 3.0 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.7 | 2.2 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.7 | 10.4 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.7 | 12.6 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.7 | 3.6 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.7 | 2.1 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.7 | 2.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.7 | 8.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.7 | 2.7 | GO:0043273 | CTPase activity(GO:0043273) |
| 0.7 | 4.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.7 | 2.0 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.7 | 3.3 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.7 | 8.5 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.6 | 41.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.6 | 3.8 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.6 | 2.5 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.6 | 4.4 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.6 | 3.8 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.6 | 12.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.6 | 3.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.6 | 3.8 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.6 | 3.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.6 | 1.9 | GO:0004912 | interleukin-3 receptor activity(GO:0004912) interleukin-5 receptor activity(GO:0004914) |
| 0.6 | 17.0 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.6 | 4.8 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.6 | 4.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.6 | 7.0 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.6 | 16.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.6 | 10.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.6 | 2.8 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.6 | 3.3 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.5 | 2.7 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.5 | 6.9 | GO:0016918 | retinal binding(GO:0016918) |
| 0.5 | 1.6 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 0.5 | 2.6 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.5 | 9.4 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.5 | 3.0 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.5 | 4.0 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.5 | 2.4 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.5 | 9.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.5 | 2.3 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.5 | 1.9 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) tetrahydrobiopterin binding(GO:0034617) |
| 0.5 | 10.7 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.4 | 13.0 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.4 | 18.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.4 | 5.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.4 | 10.1 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.4 | 25.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.4 | 13.9 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.4 | 5.6 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.4 | 6.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.4 | 8.4 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.4 | 7.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.4 | 5.8 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.4 | 5.8 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.4 | 2.9 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.4 | 31.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.4 | 4.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.4 | 1.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.4 | 5.8 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.4 | 5.4 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.4 | 27.1 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.4 | 3.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.4 | 2.6 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.4 | 1.5 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.4 | 1.1 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.4 | 2.6 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.4 | 6.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.4 | 5.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.4 | 1.4 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.4 | 2.1 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.4 | 5.0 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.4 | 2.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.3 | 2.4 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.3 | 5.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.3 | 5.5 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.3 | 3.4 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.3 | 2.7 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.3 | 2.6 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.3 | 2.0 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.3 | 2.6 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.3 | 9.4 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.3 | 1.3 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.3 | 4.4 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.3 | 8.0 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.3 | 3.0 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.3 | 1.8 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.3 | 4.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.3 | 10.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.3 | 7.4 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.3 | 4.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.3 | 10.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.3 | 1.1 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.3 | 1.4 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.3 | 3.9 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.3 | 4.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.3 | 11.7 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.3 | 1.8 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.3 | 9.7 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.2 | 24.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 5.5 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 24.6 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.2 | 6.4 | GO:0008066 | glutamate receptor activity(GO:0008066) |
| 0.2 | 0.7 | GO:0047783 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.2 | 5.1 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.2 | 2.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 5.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 12.0 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.2 | 6.7 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 8.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.2 | 2.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 1.3 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.2 | 2.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.2 | 4.8 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.2 | 1.0 | GO:0008948 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) oxaloacetate decarboxylase activity(GO:0008948) |
| 0.2 | 2.1 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.2 | 0.6 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.2 | 0.8 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.2 | 2.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.2 | 0.7 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.2 | 2.9 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.2 | 2.0 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.2 | 4.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 4.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 2.7 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.2 | 3.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.2 | 11.6 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.2 | 1.8 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 12.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 3.9 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.2 | 10.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.2 | 1.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.2 | 1.4 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 5.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 1.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.9 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 5.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 1.5 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 4.9 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 1.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 3.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 4.2 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 3.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 1.5 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 3.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.8 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.9 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 1.8 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.1 | 4.9 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 5.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.3 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 13.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 13.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 63.5 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 3.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 7.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 0.8 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 134.5 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.1 | 1.0 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 9.5 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 6.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 2.4 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 2.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.9 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 2.3 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 22.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 0.4 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.1 | 2.4 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.1 | 6.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 3.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.5 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.1 | 1.0 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.7 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.6 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 1.4 | GO:0042171 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.1 | 0.3 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 1.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.3 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 1.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 2.5 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 3.2 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 2.5 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.7 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.3 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 1.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 3.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 7.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 7.9 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 1.2 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.0 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 1.0 | 106.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.7 | 36.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.7 | 12.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.6 | 34.7 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.5 | 29.1 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.5 | 17.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.5 | 41.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.5 | 14.6 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.5 | 13.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.4 | 29.9 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.4 | 3.6 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.4 | 81.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.4 | 7.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.4 | 2.8 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.3 | 22.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.3 | 18.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.3 | 38.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.3 | 14.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.3 | 15.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 12.1 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.3 | 8.5 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.3 | 5.6 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.3 | 57.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.3 | 1.9 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.3 | 8.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.3 | 6.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.3 | 11.0 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.3 | 5.8 | ST ADRENERGIC | Adrenergic Pathway |
| 0.2 | 5.3 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.2 | 6.0 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.2 | 14.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.2 | 12.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 17.6 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.2 | 8.4 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.2 | 23.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.2 | 4.5 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.2 | 3.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.2 | 5.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 1.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.2 | 3.6 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.2 | 14.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 2.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 4.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 1.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 0.5 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 6.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 3.2 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 3.9 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 1.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 3.2 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 1.0 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 1.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 19.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 0.6 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 0.8 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 0.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 5.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 2.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 0.8 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 2.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.4 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.4 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 120.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 1.7 | 29.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 1.2 | 18.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 1.1 | 19.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.9 | 6.4 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.8 | 16.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.7 | 14.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.7 | 12.6 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.7 | 27.8 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.7 | 9.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.7 | 11.8 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.6 | 7.0 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.6 | 4.4 | REACTOME GAB1 SIGNALOSOME | Genes involved in GAB1 signalosome |
| 0.6 | 12.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.6 | 44.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.6 | 15.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.6 | 11.1 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.5 | 23.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.5 | 18.9 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.5 | 43.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.5 | 11.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.5 | 26.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.5 | 7.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.5 | 9.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.5 | 10.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.4 | 8.0 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.4 | 10.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.4 | 11.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.4 | 30.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.4 | 22.7 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.4 | 0.8 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.4 | 13.1 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.4 | 7.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.4 | 22.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.4 | 48.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.4 | 5.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.3 | 35.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.3 | 4.8 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.3 | 2.2 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.3 | 9.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.3 | 25.0 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.3 | 2.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.3 | 8.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.3 | 5.6 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.3 | 6.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.3 | 31.4 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.3 | 8.9 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.3 | 6.4 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.3 | 6.9 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.3 | 11.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.3 | 12.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.3 | 4.0 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.3 | 4.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.3 | 4.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.3 | 5.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.3 | 4.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 15.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 9.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 44.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 14.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 11.0 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.2 | 10.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 5.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 1.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.2 | 5.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 3.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 4.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 5.3 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.2 | 6.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.2 | 4.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 5.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 8.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 4.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 4.0 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 0.7 | REACTOME GABA B RECEPTOR ACTIVATION | Genes involved in GABA B receptor activation |
| 0.1 | 4.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 6.1 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 2.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 5.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 4.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 2.8 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 15.9 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 2.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.6 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 2.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.9 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 23.3 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.1 | 1.8 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 1.5 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 0.9 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 0.8 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 1.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.5 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.1 | 1.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.0 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.1 | 2.0 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 1.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.4 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 1.3 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 0.3 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.8 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.2 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |