Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
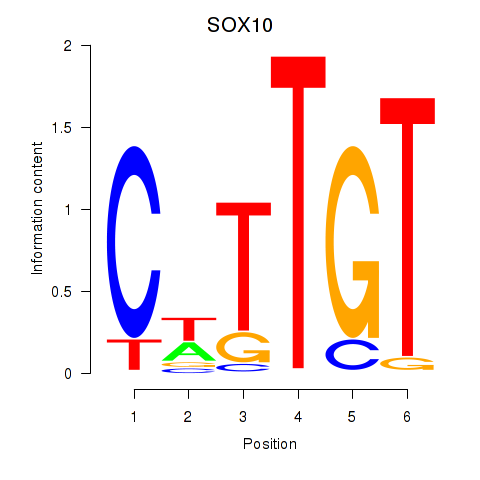
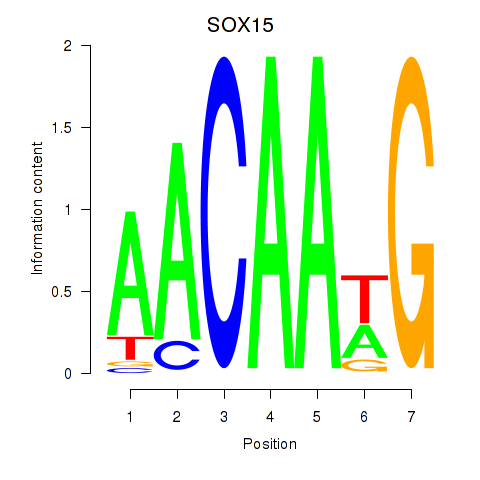
Results for SOX10_SOX15
Z-value: 0.92
Transcription factors associated with SOX10_SOX15
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX10
|
ENSG00000100146.18 | SOX10 |
|
SOX15
|
ENSG00000129194.8 | SOX15 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX15 | hg38_v1_chr17_-_7590072_7590101 | -0.54 | 8.3e-18 | Click! |
| SOX10 | hg38_v1_chr22_-_37984534_37984562 | -0.48 | 9.1e-14 | Click! |
Activity profile of SOX10_SOX15 motif
Sorted Z-values of SOX10_SOX15 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX10_SOX15
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.9 | 39.7 | GO:0032597 | B cell receptor transport within lipid bilayer(GO:0032595) B cell receptor transport into membrane raft(GO:0032597) protein transport out of membrane raft(GO:0032599) chemokine receptor transport out of membrane raft(GO:0032600) negative regulation of transforming growth factor beta3 production(GO:0032913) chemokine receptor transport within lipid bilayer(GO:0033606) |
| 5.4 | 21.6 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 4.5 | 31.7 | GO:1901297 | arterial endothelial cell fate commitment(GO:0060844) blood vessel lumenization(GO:0072554) positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 2.9 | 25.9 | GO:0007070 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 2.7 | 5.4 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 2.6 | 18.5 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 2.6 | 10.3 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 2.4 | 12.2 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 2.3 | 58.6 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 2.3 | 9.3 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 2.2 | 22.4 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 1.9 | 15.1 | GO:2000483 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) negative regulation of interleukin-8 secretion(GO:2000483) |
| 1.6 | 6.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 1.5 | 10.6 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 1.4 | 5.6 | GO:0003290 | atrial septum secundum morphogenesis(GO:0003290) |
| 1.4 | 4.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 1.3 | 14.4 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 1.3 | 21.7 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 1.2 | 5.0 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 1.2 | 4.9 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 1.1 | 9.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 1.1 | 8.6 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 1.0 | 4.2 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 1.0 | 18.7 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 1.0 | 3.0 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 1.0 | 15.0 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.9 | 4.7 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.9 | 4.6 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.9 | 9.8 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.9 | 2.7 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.9 | 8.8 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.9 | 2.6 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.9 | 5.3 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.9 | 9.6 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.8 | 1.7 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.8 | 14.1 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.8 | 3.3 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.8 | 4.9 | GO:0042997 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.8 | 5.6 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.8 | 4.0 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.8 | 12.7 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.8 | 20.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.8 | 11.7 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.8 | 4.6 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.8 | 9.2 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.8 | 5.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.7 | 3.7 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.7 | 2.2 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.7 | 19.8 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.7 | 2.1 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.7 | 2.0 | GO:1903004 | flavin adenine dinucleotide metabolic process(GO:0072387) regulation of protein K63-linked deubiquitination(GO:1903004) positive regulation of protein K63-linked deubiquitination(GO:1903006) |
| 0.7 | 16.8 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.7 | 10.0 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.6 | 11.5 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.6 | 18.4 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.6 | 13.1 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.6 | 5.6 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.6 | 4.9 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.6 | 11.0 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.6 | 5.5 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.6 | 8.3 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.6 | 7.7 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.6 | 4.1 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.6 | 4.0 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.6 | 1.7 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.6 | 7.9 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.5 | 24.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.5 | 3.6 | GO:0030421 | defecation(GO:0030421) |
| 0.5 | 2.6 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.5 | 5.5 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.5 | 2.9 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.5 | 5.0 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.4 | 5.8 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.4 | 6.5 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) |
| 0.4 | 6.0 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.4 | 7.1 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.4 | 1.7 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.4 | 3.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.4 | 5.2 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.4 | 1.6 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.4 | 1.6 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.4 | 9.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.4 | 2.7 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.4 | 6.9 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.4 | 8.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.4 | 7.2 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.4 | 7.0 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.3 | 2.4 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.3 | 5.1 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.3 | 12.8 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.3 | 4.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.3 | 2.7 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.3 | 3.9 | GO:0042407 | cristae formation(GO:0042407) |
| 0.3 | 1.9 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.3 | 2.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.3 | 1.5 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.3 | 7.9 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.3 | 6.2 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.3 | 9.5 | GO:0060065 | uterus development(GO:0060065) |
| 0.3 | 0.3 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.3 | 2.7 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.3 | 9.4 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.3 | 0.5 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.3 | 2.8 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.3 | 0.8 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.2 | 7.3 | GO:0072662 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.2 | 1.4 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.2 | 0.8 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.2 | 0.8 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.2 | 3.6 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.2 | 2.6 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.2 | 2.4 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.2 | 1.2 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.2 | 12.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.2 | 1.6 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.2 | 1.1 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.2 | 9.0 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.2 | 2.7 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 0.4 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.2 | 1.9 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.2 | 20.1 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.2 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.2 | 0.5 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.2 | 21.7 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 1.1 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.1 | 0.6 | GO:0001916 | positive regulation of T cell mediated cytotoxicity(GO:0001916) |
| 0.1 | 0.3 | GO:0002715 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell mediated immunity(GO:0002715) negative regulation of natural killer cell mediated immunity(GO:0002716) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.1 | 1.1 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.1 | 0.4 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.1 | 8.9 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.1 | 1.3 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 10.1 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.1 | 3.1 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.1 | 0.7 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 2.4 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 3.9 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.1 | 1.3 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.1 | 3.3 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.1 | 15.3 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.1 | 1.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 5.5 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.1 | 4.2 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.1 | 6.9 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.1 | 1.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 3.0 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.1 | 3.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 14.3 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 2.1 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.1 | 1.0 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 1.3 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.1 | 0.3 | GO:0021836 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) negative regulation of mammary gland epithelial cell proliferation(GO:0033600) regulation of negative chemotaxis(GO:0050923) |
| 0.1 | 2.5 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 | 2.6 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 0.9 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.1 | 1.3 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 1.2 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.1 | 4.7 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 4.2 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.1 | 5.3 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.1 | 7.1 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 5.9 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.1 | 5.1 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.1 | 0.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.7 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.2 | GO:0030862 | positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.0 | 2.6 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 1.1 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.3 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.4 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 3.1 | GO:0034766 | negative regulation of ion transmembrane transport(GO:0034766) |
| 0.0 | 1.1 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.6 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 6.0 | GO:0043279 | response to alkaloid(GO:0043279) |
| 0.0 | 0.2 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.0 | 6.9 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 0.4 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 2.9 | GO:0006275 | regulation of DNA replication(GO:0006275) |
| 0.0 | 0.5 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 11.5 | GO:0042493 | response to drug(GO:0042493) |
| 0.0 | 15.6 | GO:0043312 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 1.8 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 0.7 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 2.9 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 3.0 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.0 | 0.1 | GO:0034370 | triglyceride-rich lipoprotein particle remodeling(GO:0034370) very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.4 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 2.2 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.5 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 1.0 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.0 | 0.3 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.6 | 58.6 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 5.1 | 15.3 | GO:0008623 | CHRAC(GO:0008623) |
| 3.8 | 34.3 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 2.5 | 22.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 2.1 | 6.3 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 1.9 | 18.7 | GO:0070852 | cell body fiber(GO:0070852) |
| 1.7 | 39.7 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 1.6 | 8.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 1.4 | 18.7 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 1.4 | 8.6 | GO:0097422 | tubular endosome(GO:0097422) |
| 1.3 | 9.3 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 1.3 | 20.3 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 1.0 | 3.0 | GO:0044393 | microspike(GO:0044393) |
| 0.9 | 32.7 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.9 | 7.4 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.9 | 7.9 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.8 | 9.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.8 | 10.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.8 | 18.2 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.8 | 9.6 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.8 | 5.5 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.6 | 3.9 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.6 | 5.0 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.6 | 5.7 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.5 | 13.8 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.5 | 2.0 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.4 | 4.6 | GO:0005638 | lamin filament(GO:0005638) |
| 0.4 | 5.4 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.3 | 1.6 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.3 | 5.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.3 | 5.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.3 | 1.7 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.3 | 4.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.2 | 2.6 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.2 | 0.9 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.2 | 1.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.2 | 4.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.2 | 2.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 1.3 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.2 | 13.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 2.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.2 | 1.2 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.2 | 3.0 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 2.1 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 0.7 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 14.6 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 23.2 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 6.5 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 5.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 11.9 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 8.1 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 4.0 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 14.7 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 16.5 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 1.1 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.3 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.1 | 12.7 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.1 | 1.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 2.2 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.1 | 9.6 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 3.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 1.6 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 0.8 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 2.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 2.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 3.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 19.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 6.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 3.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 11.9 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.1 | 3.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 3.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 13.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 22.0 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 1.8 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 4.0 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 3.4 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 14.0 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 6.3 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 2.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 2.5 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 3.4 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 9.4 | GO:0005694 | chromosome(GO:0005694) |
| 0.0 | 10.4 | GO:0030529 | intracellular ribonucleoprotein complex(GO:0030529) |
| 0.0 | 1.7 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.1 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 2.2 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.2 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.6 | 58.6 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 7.2 | 21.6 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 4.4 | 13.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 4.3 | 13.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 4.3 | 12.9 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 2.0 | 31.7 | GO:0000150 | recombinase activity(GO:0000150) |
| 1.9 | 9.6 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 1.8 | 9.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 1.8 | 7.2 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 1.4 | 25.9 | GO:0030957 | Tat protein binding(GO:0030957) |
| 1.3 | 39.6 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 1.2 | 3.7 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 1.1 | 5.7 | GO:0017161 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 1.1 | 5.4 | GO:0004515 | nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 1.0 | 39.7 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 1.0 | 5.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 1.0 | 5.8 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.9 | 8.8 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.9 | 5.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.9 | 18.4 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.8 | 5.6 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.8 | 20.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.7 | 2.2 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.7 | 11.5 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.7 | 2.9 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.7 | 5.4 | GO:0070699 | activin receptor binding(GO:0070697) type II activin receptor binding(GO:0070699) |
| 0.7 | 11.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.6 | 12.6 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.6 | 4.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.6 | 18.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.5 | 19.6 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.5 | 24.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.5 | 4.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.5 | 14.5 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.5 | 4.0 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.5 | 18.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.5 | 2.8 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.5 | 7.8 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.5 | 5.5 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.5 | 15.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.4 | 9.8 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.4 | 17.4 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.4 | 7.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.4 | 2.9 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.4 | 2.9 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.4 | 24.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.4 | 16.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.4 | 1.9 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.3 | 1.7 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.3 | 2.1 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.3 | 5.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.3 | 8.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.3 | 4.7 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.3 | 1.6 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.3 | 6.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.3 | 5.6 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.2 | 6.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 0.5 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.2 | 10.0 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.2 | 10.1 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.2 | 1.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.2 | 2.7 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.2 | 7.7 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.2 | 15.3 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.2 | 2.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 5.1 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.2 | 1.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 5.5 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.2 | 3.5 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 14.8 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.2 | 2.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.2 | 1.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 3.3 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 5.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 7.3 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 29.6 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 15.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 4.8 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 1.7 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 4.0 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 1.8 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 48.9 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 0.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 2.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 8.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.4 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 5.3 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 2.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 24.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 4.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 2.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 2.6 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 5.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 0.7 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.3 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 1.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 9.2 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 0.3 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.1 | 3.0 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 0.6 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 4.0 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) NAD binding(GO:0051287) |
| 0.1 | 0.4 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 8.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 2.3 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 0.6 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.7 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 1.8 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 3.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:0031782 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 7.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 2.1 | GO:0032934 | sterol binding(GO:0032934) |
| 0.0 | 14.2 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.0 | 0.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 3.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 2.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 3.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.7 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.6 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 23.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.9 | 25.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.6 | 35.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.6 | 31.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.5 | 72.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.5 | 10.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.5 | 39.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.4 | 7.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.3 | 12.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.3 | 9.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 36.1 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.3 | 17.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.3 | 3.9 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.2 | 7.6 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.2 | 14.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 4.7 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.2 | 7.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.2 | 7.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 5.0 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 8.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 5.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 18.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 6.8 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 2.7 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 4.3 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.1 | 5.3 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 9.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 2.7 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 3.0 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 5.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 5.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 4.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 1.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 2.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 2.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 8.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.7 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 34.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 1.8 | 58.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 1.7 | 20.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 1.5 | 8.8 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 1.1 | 18.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.9 | 18.9 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.8 | 15.6 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.7 | 38.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.7 | 21.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.5 | 10.9 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.5 | 10.0 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.4 | 13.1 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.4 | 9.2 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.3 | 9.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.3 | 8.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.3 | 4.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.3 | 4.8 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.3 | 19.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.3 | 6.0 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.3 | 20.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.2 | 6.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.2 | 34.8 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.2 | 3.9 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 4.9 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.2 | 6.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 2.7 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.2 | 1.7 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 3.0 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 4.0 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 19.3 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 1.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 2.9 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 2.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 5.7 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 7.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 13.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 5.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 7.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 2.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 2.4 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 0.6 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 3.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.6 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 10.1 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.1 | 5.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 1.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 5.5 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.1 | 0.5 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 2.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 6.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.0 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 2.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |