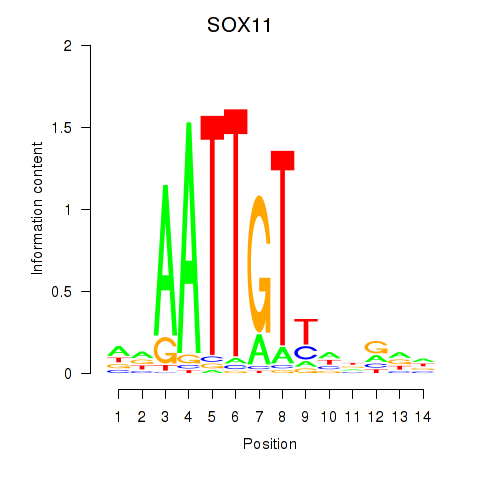
|
chr20_+_11892493
Show fit
|
10.52 |
ENST00000422390.5
ENST00000618918.4
|
BTBD3
|
BTB domain containing 3
|
|
chr15_+_80441229
Show fit
|
10.04 |
ENST00000533983.5
ENST00000527771.5
ENST00000525103.1
|
ARNT2
|
aryl hydrocarbon receptor nuclear translocator 2
|
|
chr5_+_72107453
Show fit
|
7.97 |
ENST00000296755.12
ENST00000511641.2
|
MAP1B
|
microtubule associated protein 1B
|
|
chr4_-_89838289
Show fit
|
7.14 |
ENST00000336904.7
|
SNCA
|
synuclein alpha
|
|
chr13_-_44474296
Show fit
|
6.68 |
ENST00000611198.4
|
TSC22D1
|
TSC22 domain family member 1
|
|
chr7_-_93890160
Show fit
|
6.46 |
ENST00000451238.1
|
TFPI2
|
tissue factor pathway inhibitor 2
|
|
chrX_-_13817027
Show fit
|
6.42 |
ENST00000493677.5
ENST00000355135.6
ENST00000316715.9
|
GPM6B
|
glycoprotein M6B
|
|
chr9_-_14300231
Show fit
|
5.55 |
ENST00000636735.1
|
NFIB
|
nuclear factor I B
|
|
chr17_-_10114546
Show fit
|
4.76 |
ENST00000323816.8
|
GAS7
|
growth arrest specific 7
|
|
chr8_-_13276491
Show fit
|
4.18 |
ENST00000512044.6
|
DLC1
|
DLC1 Rho GTPase activating protein
|
|
chr7_+_99325857
Show fit
|
4.01 |
ENST00000638617.1
ENST00000262942.10
|
ENSG00000284292.1
ARPC1A
|
novel protein, ARPC1A and ARPC1B readthrough
actin related protein 2/3 complex subunit 1A
|
|
chr1_+_204870831
Show fit
|
3.43 |
ENST00000404076.5
ENST00000539706.6
|
NFASC
|
neurofascin
|
|
chr15_+_83447328
Show fit
|
3.28 |
ENST00000427482.7
|
SH3GL3
|
SH3 domain containing GRB2 like 3, endophilin A3
|
|
chr7_-_83162899
Show fit
|
3.21 |
ENST00000423517.6
|
PCLO
|
piccolo presynaptic cytomatrix protein
|
|
chr2_+_11534039
Show fit
|
3.21 |
ENST00000381486.7
|
GREB1
|
growth regulating estrogen receptor binding 1
|
|
chr2_-_224402097
Show fit
|
3.11 |
ENST00000409685.4
|
FAM124B
|
family with sequence similarity 124 member B
|
|
chr15_+_83447411
Show fit
|
3.00 |
ENST00000324537.5
|
SH3GL3
|
SH3 domain containing GRB2 like 3, endophilin A3
|
|
chr17_-_68955332
Show fit
|
2.98 |
ENST00000269080.6
ENST00000615593.4
ENST00000586539.6
ENST00000430352.6
|
ABCA8
|
ATP binding cassette subfamily A member 8
|
|
chr2_-_2324323
Show fit
|
2.72 |
ENST00000648339.1
ENST00000647694.1
|
MYT1L
|
myelin transcription factor 1 like
|
|
chr8_-_53839846
Show fit
|
2.64 |
ENST00000520188.5
|
ATP6V1H
|
ATPase H+ transporting V1 subunit H
|
|
chr18_+_7754959
Show fit
|
2.62 |
ENST00000400053.8
|
PTPRM
|
protein tyrosine phosphatase receptor type M
|
|
chr2_-_224401994
Show fit
|
2.61 |
ENST00000389874.3
|
FAM124B
|
family with sequence similarity 124 member B
|
|
chr5_+_174045673
Show fit
|
2.56 |
ENST00000303177.8
ENST00000519867.5
|
NSG2
|
neuronal vesicle trafficking associated 2
|
|
chr10_-_93482194
Show fit
|
2.50 |
ENST00000358334.9
ENST00000371488.3
|
MYOF
|
myoferlin
|
|
chr2_+_137964279
Show fit
|
2.49 |
ENST00000329366.8
|
HNMT
|
histamine N-methyltransferase
|
|
chr10_-_93482326
Show fit
|
2.49 |
ENST00000359263.9
|
MYOF
|
myoferlin
|
|
chr13_-_29850605
Show fit
|
2.44 |
ENST00000380680.5
|
UBL3
|
ubiquitin like 3
|
|
chr5_+_140868945
Show fit
|
2.42 |
ENST00000398640.7
|
PCDHA11
|
protocadherin alpha 11
|
|
chr2_-_189179754
Show fit
|
2.28 |
ENST00000374866.9
ENST00000618828.1
|
COL5A2
|
collagen type V alpha 2 chain
|
|
chr6_-_47309898
Show fit
|
2.11 |
ENST00000296861.2
|
TNFRSF21
|
TNF receptor superfamily member 21
|
|
chr5_+_140834230
Show fit
|
2.08 |
ENST00000356878.5
ENST00000525929.2
|
PCDHA7
|
protocadherin alpha 7
|
|
chr10_-_125816596
Show fit
|
2.04 |
ENST00000368786.5
|
UROS
|
uroporphyrinogen III synthase
|
|
chr5_+_149141817
Show fit
|
2.00 |
ENST00000504238.5
|
ABLIM3
|
actin binding LIM protein family member 3
|
|
chr2_+_69013414
Show fit
|
1.99 |
ENST00000681816.1
ENST00000482235.2
|
ANTXR1
|
ANTXR cell adhesion molecule 1
|
|
chr6_+_30617825
Show fit
|
1.98 |
ENST00000259873.5
|
MRPS18B
|
mitochondrial ribosomal protein S18B
|
|
chr17_+_6444441
Show fit
|
1.89 |
ENST00000250056.12
ENST00000571373.5
ENST00000570337.6
ENST00000572595.6
ENST00000572447.6
ENST00000576056.5
|
PIMREG
|
PICALM interacting mitotic regulator
|
|
chr1_+_65264694
Show fit
|
1.87 |
ENST00000263441.11
ENST00000395325.7
|
DNAJC6
|
DnaJ heat shock protein family (Hsp40) member C6
|
|
chr6_-_11382247
Show fit
|
1.83 |
ENST00000397378.7
ENST00000513989.5
ENST00000508546.5
ENST00000504387.5
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr2_+_69013379
Show fit
|
1.75 |
ENST00000409349.7
|
ANTXR1
|
ANTXR cell adhesion molecule 1
|
|
chr1_+_205042960
Show fit
|
1.74 |
ENST00000638378.1
|
CNTN2
|
contactin 2
|
|
chr15_-_64381431
Show fit
|
1.72 |
ENST00000558008.3
ENST00000300035.9
ENST00000559519.5
ENST00000380258.6
|
PCLAF
|
PCNA clamp associated factor
|
|
chr16_+_67227105
Show fit
|
1.67 |
ENST00000563953.5
ENST00000304800.14
ENST00000565201.1
|
TMEM208
|
transmembrane protein 208
|
|
chr5_+_140875299
Show fit
|
1.65 |
ENST00000613593.1
ENST00000398631.3
|
PCDHA12
|
protocadherin alpha 12
|
|
chr22_-_30246739
Show fit
|
1.62 |
ENST00000403987.3
ENST00000249075.4
|
LIF
|
LIF interleukin 6 family cytokine
|
|
chr3_-_123980727
Show fit
|
1.55 |
ENST00000620893.4
|
ROPN1
|
rhophilin associated tail protein 1
|
|
chr15_+_57219411
Show fit
|
1.48 |
ENST00000543579.5
ENST00000537840.5
ENST00000343827.7
|
TCF12
|
transcription factor 12
|
|
chr17_-_81937221
Show fit
|
1.48 |
ENST00000402252.6
ENST00000583564.5
ENST00000585244.1
ENST00000337943.9
ENST00000579698.5
|
PYCR1
|
pyrroline-5-carboxylate reductase 1
|
|
chr1_-_112935984
Show fit
|
1.40 |
ENST00000443580.6
|
SLC16A1
|
solute carrier family 16 member 1
|
|
chr18_+_11752041
Show fit
|
1.33 |
ENST00000423027.8
|
GNAL
|
G protein subunit alpha L
|
|
chr7_-_96709780
Show fit
|
1.31 |
ENST00000623693.3
ENST00000623498.3
ENST00000488005.5
ENST00000413065.5
|
SEM1
|
SEM1 26S proteasome complex subunit
|
|
chr22_-_28306645
Show fit
|
1.28 |
ENST00000612946.4
|
TTC28
|
tetratricopeptide repeat domain 28
|
|
chr17_-_36017953
Show fit
|
1.28 |
ENST00000612516.4
ENST00000615050.2
|
CCL23
|
C-C motif chemokine ligand 23
|
|
chr17_-_81937277
Show fit
|
1.27 |
ENST00000405481.8
ENST00000329875.13
ENST00000585215.5
|
PYCR1
|
pyrroline-5-carboxylate reductase 1
|
|
chr5_-_160852200
Show fit
|
1.22 |
ENST00000327245.10
|
ATP10B
|
ATPase phospholipid transporting 10B (putative)
|
|
chr2_+_137964446
Show fit
|
1.22 |
ENST00000280096.5
ENST00000280097.5
|
HNMT
|
histamine N-methyltransferase
|
|
chr8_+_104339796
Show fit
|
1.15 |
ENST00000622554.1
ENST00000297581.2
|
DCSTAMP
|
dendrocyte expressed seven transmembrane protein
|
|
chr2_+_73828916
Show fit
|
1.15 |
ENST00000339566.7
ENST00000683349.1
ENST00000682157.1
ENST00000682851.1
ENST00000394070.7
ENST00000409707.6
ENST00000684585.1
ENST00000683434.1
ENST00000682352.1
ENST00000424659.6
ENST00000536064.2
ENST00000684355.1
ENST00000682998.1
ENST00000682799.1
ENST00000394073.6
ENST00000682848.1
ENST00000683877.1
ENST00000682351.1
ENST00000683818.1
ENST00000683391.1
ENST00000682558.1
|
STAMBP
|
STAM binding protein
|
|
chr3_-_123991352
Show fit
|
1.13 |
ENST00000184183.8
|
ROPN1
|
rhophilin associated tail protein 1
|
|
chr8_+_9555900
Show fit
|
1.12 |
ENST00000310430.11
ENST00000520408.5
ENST00000522110.1
|
TNKS
|
tankyrase
|
|
chr1_-_145910031
Show fit
|
1.03 |
ENST00000369304.8
|
ITGA10
|
integrin subunit alpha 10
|
|
chr17_-_36001549
Show fit
|
0.95 |
ENST00000617897.2
|
CCL15
|
C-C motif chemokine ligand 15
|
|
chr18_+_58196736
Show fit
|
0.94 |
ENST00000675221.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase
|
|
chr9_-_92404559
Show fit
|
0.92 |
ENST00000262551.8
ENST00000375561.10
|
OGN
|
osteoglycin
|
|
chr15_+_49155750
Show fit
|
0.90 |
ENST00000327171.7
ENST00000560654.5
|
GALK2
|
galactokinase 2
|
|
chr11_-_118176576
Show fit
|
0.90 |
ENST00000278947.6
|
SCN2B
|
sodium voltage-gated channel beta subunit 2
|
|
chr14_-_36320582
Show fit
|
0.90 |
ENST00000604336.5
ENST00000359527.11
ENST00000621657.4
ENST00000603139.1
ENST00000318473.11
ENST00000416007.9
|
MBIP
|
MAP3K12 binding inhibitory protein 1
|
|
chr2_-_175181663
Show fit
|
0.88 |
ENST00000392541.3
ENST00000284727.9
ENST00000409194.5
|
ATP5MC3
|
ATP synthase membrane subunit c locus 3
|
|
chr3_-_125520165
Show fit
|
0.86 |
ENST00000251775.9
|
SNX4
|
sorting nexin 4
|
|
chr18_+_48539017
Show fit
|
0.84 |
ENST00000256413.8
|
CTIF
|
cap binding complex dependent translation initiation factor
|
|
chrX_-_21758021
Show fit
|
0.83 |
ENST00000646008.1
|
SMPX
|
small muscle protein X-linked
|
|
chr2_-_177263522
Show fit
|
0.83 |
ENST00000448782.5
ENST00000446151.6
|
NFE2L2
|
nuclear factor, erythroid 2 like 2
|
|
chr1_-_145910066
Show fit
|
0.80 |
ENST00000539363.2
|
ITGA10
|
integrin subunit alpha 10
|
|
chr8_-_85341659
Show fit
|
0.79 |
ENST00000522389.5
|
CA1
|
carbonic anhydrase 1
|
|
chr21_+_5011799
Show fit
|
0.79 |
ENST00000624081.1
|
ENSG00000279493.1
|
novel protein, similar to DNA (cytosine-5-)-methyltransferase 3-like DNMT3L
|
|
chr2_+_102311546
Show fit
|
0.78 |
ENST00000233954.6
ENST00000447231.5
|
IL1RL1
|
interleukin 1 receptor like 1
|
|
chr7_+_70596078
Show fit
|
0.78 |
ENST00000644506.1
|
AUTS2
|
activator of transcription and developmental regulator AUTS2
|
|
chr11_+_22666604
Show fit
|
0.75 |
ENST00000454584.6
|
GAS2
|
growth arrest specific 2
|
|
chr4_+_71339014
Show fit
|
0.68 |
ENST00000340595.4
|
SLC4A4
|
solute carrier family 4 member 4
|
|
chr9_-_21141832
Show fit
|
0.68 |
ENST00000380229.4
|
IFNW1
|
interferon omega 1
|
|
chr2_+_102311502
Show fit
|
0.67 |
ENST00000404917.6
ENST00000410040.5
|
IL1RL1
IL18R1
|
interleukin 1 receptor like 1
interleukin 18 receptor 1
|
|
chr8_+_103819244
Show fit
|
0.67 |
ENST00000262231.14
ENST00000507740.5
ENST00000408894.6
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr14_+_102362931
Show fit
|
0.65 |
ENST00000359520.12
|
TECPR2
|
tectonin beta-propeller repeat containing 2
|
|
chr16_+_482507
Show fit
|
0.62 |
ENST00000412256.1
|
RAB11FIP3
|
RAB11 family interacting protein 3
|
|
chr12_-_10802627
Show fit
|
0.60 |
ENST00000240687.2
|
TAS2R7
|
taste 2 receptor member 7
|
|
chr9_+_73151833
Show fit
|
0.60 |
ENST00000456643.5
ENST00000257497.11
ENST00000415424.5
|
ANXA1
|
annexin A1
|
|
chr10_+_80408485
Show fit
|
0.59 |
ENST00000615554.4
ENST00000372185.5
|
PRXL2A
|
peroxiredoxin like 2A
|
|
chr6_+_52423680
Show fit
|
0.56 |
ENST00000538167.2
|
EFHC1
|
EF-hand domain containing 1
|
|
chr6_+_26183750
Show fit
|
0.54 |
ENST00000614097.3
|
H2BC6
|
H2B clustered histone 6
|
|
chr6_-_11778781
Show fit
|
0.54 |
ENST00000414691.8
ENST00000229583.9
|
ADTRP
|
androgen dependent TFPI regulating protein
|
|
chr20_-_7940444
Show fit
|
0.53 |
ENST00000378789.4
|
HAO1
|
hydroxyacid oxidase 1
|
|
chrX_+_30243715
Show fit
|
0.53 |
ENST00000378981.8
ENST00000397550.6
|
MAGEB1
|
MAGE family member B1
|
|
chr5_+_136160986
Show fit
|
0.51 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5
|
|
chr6_-_46080332
Show fit
|
0.51 |
ENST00000185206.12
|
CLIC5
|
chloride intracellular channel 5
|
|
chrX_-_7927701
Show fit
|
0.50 |
ENST00000537427.5
ENST00000444736.5
ENST00000442940.1
|
PNPLA4
|
patatin like phospholipase domain containing 4
|
|
chr8_+_24384275
Show fit
|
0.48 |
ENST00000256412.8
|
ADAMDEC1
|
ADAM like decysin 1
|
|
chr20_+_33237712
Show fit
|
0.46 |
ENST00000618484.1
|
BPIFA1
|
BPI fold containing family A member 1
|
|
chr7_-_122995700
Show fit
|
0.46 |
ENST00000249284.3
|
TAS2R16
|
taste 2 receptor member 16
|
|
chr11_+_62208665
Show fit
|
0.45 |
ENST00000244930.6
|
SCGB2A1
|
secretoglobin family 2A member 1
|
|
chr2_+_108621260
Show fit
|
0.43 |
ENST00000409441.5
|
LIMS1
|
LIM zinc finger domain containing 1
|
|
chr8_-_85341705
Show fit
|
0.43 |
ENST00000517618.5
|
CA1
|
carbonic anhydrase 1
|
|
chr16_+_19067989
Show fit
|
0.42 |
ENST00000569127.1
|
COQ7
|
coenzyme Q7, hydroxylase
|
|
chr12_-_81369348
Show fit
|
0.40 |
ENST00000548670.1
ENST00000541017.5
ENST00000541570.6
ENST00000553058.5
|
PPFIA2
|
PTPRF interacting protein alpha 2
|
|
chrX_-_21758097
Show fit
|
0.39 |
ENST00000379494.4
|
SMPX
|
small muscle protein X-linked
|
|
chr4_-_158159657
Show fit
|
0.38 |
ENST00000590648.5
|
GASK1B
|
golgi associated kinase 1B
|
|
chr1_-_24143112
Show fit
|
0.37 |
ENST00000270800.2
|
IL22RA1
|
interleukin 22 receptor subunit alpha 1
|
|
chr7_+_142111739
Show fit
|
0.37 |
ENST00000550469.6
ENST00000477922.3
|
MGAM2
|
maltase-glucoamylase 2 (putative)
|
|
chr8_-_25424260
Show fit
|
0.35 |
ENST00000421054.7
|
GNRH1
|
gonadotropin releasing hormone 1
|
|
chr1_-_150697128
Show fit
|
0.33 |
ENST00000427665.1
ENST00000271732.8
|
GOLPH3L
|
golgi phosphoprotein 3 like
|
|
chr3_-_46085817
Show fit
|
0.33 |
ENST00000683768.1
|
XCR1
|
X-C motif chemokine receptor 1
|
|
chr1_-_92792396
Show fit
|
0.32 |
ENST00000370331.5
ENST00000540033.2
|
EVI5
|
ecotropic viral integration site 5
|
|
chr8_-_71361860
Show fit
|
0.31 |
ENST00000303824.11
ENST00000645451.1
|
EYA1
|
EYA transcriptional coactivator and phosphatase 1
|
|
chr8_-_71362054
Show fit
|
0.31 |
ENST00000340726.8
|
EYA1
|
EYA transcriptional coactivator and phosphatase 1
|
|
chrX_-_32155462
Show fit
|
0.29 |
ENST00000359836.5
ENST00000378707.7
ENST00000541735.5
ENST00000684130.1
ENST00000682238.1
ENST00000620040.5
ENST00000474231.5
|
DMD
|
dystrophin
|
|
chr1_+_50103903
Show fit
|
0.27 |
ENST00000371827.5
|
ELAVL4
|
ELAV like RNA binding protein 4
|
|
chr18_+_24460655
Show fit
|
0.26 |
ENST00000426880.2
|
HRH4
|
histamine receptor H4
|
|
chr4_+_168092530
Show fit
|
0.25 |
ENST00000359299.8
|
ANXA10
|
annexin A10
|
|
chr13_-_98977975
Show fit
|
0.24 |
ENST00000376460.5
|
DOCK9
|
dedicator of cytokinesis 9
|
|
chr9_+_27109200
Show fit
|
0.24 |
ENST00000380036.10
|
TEK
|
TEK receptor tyrosine kinase
|
|
chr17_-_10373002
Show fit
|
0.22 |
ENST00000252172.9
ENST00000418404.8
|
MYH13
|
myosin heavy chain 13
|
|
chr6_-_49636832
Show fit
|
0.21 |
ENST00000371175.10
ENST00000646272.1
ENST00000646939.1
ENST00000618248.3
ENST00000229810.9
ENST00000646963.1
|
RHAG
|
Rh associated glycoprotein
|
|
chrX_+_102937272
Show fit
|
0.21 |
ENST00000218249.7
|
RAB40AL
|
RAB40A like
|
|
chr5_+_170353480
Show fit
|
0.20 |
ENST00000377360.8
|
KCNIP1
|
potassium voltage-gated channel interacting protein 1
|
|
chr8_+_41529212
Show fit
|
0.20 |
ENST00000520710.5
ENST00000518671.5
|
GINS4
|
GINS complex subunit 4
|
|
chr12_+_50400809
Show fit
|
0.18 |
ENST00000293618.12
ENST00000429001.7
ENST00000398473.7
ENST00000548174.5
ENST00000548697.5
ENST00000548993.5
ENST00000614335.4
ENST00000522085.5
ENST00000615080.4
ENST00000518444.5
ENST00000551886.5
ENST00000523389.5
ENST00000518561.5
ENST00000347328.9
ENST00000550260.1
|
LARP4
|
La ribonucleoprotein 4
|
|
chr1_-_110519175
Show fit
|
0.17 |
ENST00000369771.4
|
KCNA10
|
potassium voltage-gated channel subfamily A member 10
|
|
chr20_-_33720221
Show fit
|
0.14 |
ENST00000409299.8
ENST00000217398.3
ENST00000344022.7
|
PXMP4
|
peroxisomal membrane protein 4
|
|
chr14_+_22524325
Show fit
|
0.13 |
ENST00000390517.1
|
TRAJ20
|
T cell receptor alpha joining 20
|
|
chr5_+_108747879
Show fit
|
0.13 |
ENST00000281092.9
|
FER
|
FER tyrosine kinase
|
|
chr8_+_41529242
Show fit
|
0.12 |
ENST00000523277.6
ENST00000276533.4
|
GINS4
|
GINS complex subunit 4
|
|
chr4_+_87832917
Show fit
|
0.12 |
ENST00000395102.8
ENST00000497649.6
ENST00000540395.1
ENST00000560249.5
ENST00000511670.5
ENST00000361056.3
|
MEPE
|
matrix extracellular phosphoglycoprotein
|
|
chr9_+_27109135
Show fit
|
0.10 |
ENST00000519097.5
ENST00000615002.4
|
TEK
|
TEK receptor tyrosine kinase
|
|
chr11_+_57657736
Show fit
|
0.09 |
ENST00000529773.2
ENST00000533905.1
ENST00000525602.1
ENST00000533682.2
ENST00000302731.4
|
CLP1
|
cleavage factor polyribonucleotide kinase subunit 1
|
|
chr10_+_116591010
Show fit
|
0.09 |
ENST00000530319.5
ENST00000527980.5
ENST00000471549.5
ENST00000534537.5
|
PNLIPRP1
|
pancreatic lipase related protein 1
|
|
chr19_+_15728024
Show fit
|
0.07 |
ENST00000305899.5
|
OR10H2
|
olfactory receptor family 10 subfamily H member 2
|
|
chr14_-_52069039
Show fit
|
0.07 |
ENST00000216286.10
|
NID2
|
nidogen 2
|
|
chr18_+_24460630
Show fit
|
0.05 |
ENST00000256906.5
|
HRH4
|
histamine receptor H4
|
|
chr16_+_19067893
Show fit
|
0.02 |
ENST00000544894.6
ENST00000561858.5
|
COQ7
|
coenzyme Q7, hydroxylase
|