Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
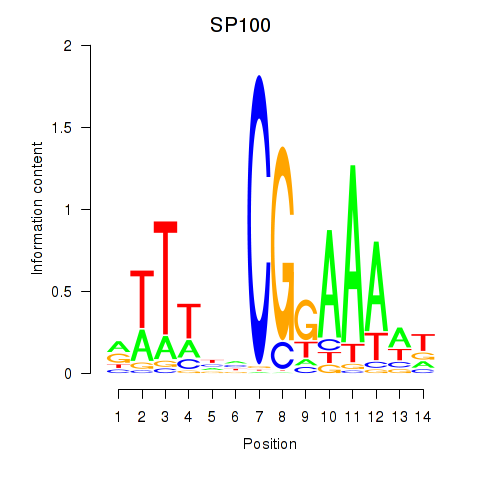
Results for SP100
Z-value: 1.44
Transcription factors associated with SP100
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SP100
|
ENSG00000067066.17 | SP100 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SP100 | hg38_v1_chr2_+_230416156_230416186 | 0.31 | 4.1e-06 | Click! |
Activity profile of SP100 motif
Sorted Z-values of SP100 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SP100
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.2 | 48.9 | GO:0002188 | translation reinitiation(GO:0002188) |
| 7.1 | 21.4 | GO:0006272 | leading strand elongation(GO:0006272) |
| 6.1 | 18.4 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 5.9 | 17.6 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 5.8 | 23.0 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 5.2 | 15.5 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 5.1 | 20.2 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 5.0 | 25.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 5.0 | 14.9 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 4.8 | 23.8 | GO:0009149 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
| 4.7 | 23.6 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 4.7 | 61.0 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 4.2 | 12.7 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 4.2 | 21.0 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 4.2 | 12.5 | GO:1904897 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 4.2 | 70.9 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 4.1 | 12.3 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 4.1 | 16.3 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 3.9 | 11.8 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 3.9 | 11.7 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 3.8 | 15.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 3.7 | 11.1 | GO:0051793 | medium-chain fatty acid catabolic process(GO:0051793) |
| 3.4 | 10.2 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 3.3 | 13.4 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 3.2 | 9.7 | GO:1901388 | regulation of transforming growth factor beta activation(GO:1901388) negative regulation of transforming growth factor beta activation(GO:1901389) |
| 3.2 | 12.8 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 3.2 | 6.3 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 3.1 | 12.3 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 3.0 | 9.0 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 3.0 | 11.9 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 2.8 | 13.9 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 2.8 | 11.0 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 2.7 | 11.0 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 2.6 | 18.4 | GO:0015866 | ADP transport(GO:0015866) |
| 2.5 | 12.3 | GO:1901662 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 2.5 | 7.4 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 2.4 | 24.4 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 2.3 | 30.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 2.3 | 18.6 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 2.3 | 37.0 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 2.3 | 32.3 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 2.3 | 16.0 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 2.3 | 15.9 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 2.1 | 12.9 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 2.1 | 8.4 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 2.1 | 16.6 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 2.0 | 14.3 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 2.0 | 10.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 2.0 | 12.0 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 2.0 | 19.9 | GO:0045176 | apical protein localization(GO:0045176) |
| 1.9 | 5.7 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 1.9 | 20.9 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 1.9 | 55.0 | GO:0051084 | 'de novo' posttranslational protein folding(GO:0051084) |
| 1.8 | 3.6 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 1.7 | 7.0 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 1.7 | 10.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 1.6 | 21.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 1.6 | 16.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 1.6 | 14.6 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 1.6 | 22.0 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 1.6 | 10.9 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 1.5 | 10.8 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 1.5 | 10.4 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902956) negative regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902957) |
| 1.4 | 6.9 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 1.4 | 10.8 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 1.3 | 4.0 | GO:0035425 | autocrine signaling(GO:0035425) |
| 1.3 | 15.9 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 1.3 | 5.3 | GO:0090164 | protein retention in Golgi apparatus(GO:0045053) asymmetric Golgi ribbon formation(GO:0090164) |
| 1.3 | 13.1 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 1.3 | 16.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 1.3 | 8.8 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 1.3 | 11.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 1.2 | 8.6 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 1.2 | 8.6 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 1.2 | 3.7 | GO:0000472 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 1.2 | 3.6 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 1.2 | 3.5 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 1.2 | 7.0 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 1.1 | 113.0 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 1.1 | 4.4 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 1.0 | 12.5 | GO:1904871 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 1.0 | 12.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 1.0 | 3.9 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.9 | 5.5 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.9 | 8.0 | GO:0046070 | dGTP metabolic process(GO:0046070) |
| 0.9 | 16.9 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.9 | 5.3 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.9 | 10.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.8 | 3.3 | GO:0006404 | RNA import into nucleus(GO:0006404) |
| 0.8 | 10.8 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.8 | 2.5 | GO:0036233 | glycine import(GO:0036233) |
| 0.8 | 9.0 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.8 | 3.2 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.8 | 5.4 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.8 | 9.9 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.8 | 7.5 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.7 | 2.2 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.7 | 27.1 | GO:0033522 | histone H2A ubiquitination(GO:0033522) |
| 0.7 | 5.1 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.7 | 21.8 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.7 | 10.9 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.7 | 1.4 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.7 | 2.1 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.7 | 30.9 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.7 | 4.1 | GO:0031438 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.7 | 20.0 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.7 | 13.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.7 | 3.3 | GO:0071964 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.7 | 3.3 | GO:0001315 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.7 | 11.1 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.6 | 2.5 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.6 | 17.2 | GO:0060148 | positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.6 | 2.4 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.6 | 4.8 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.6 | 17.3 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.6 | 1.7 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.6 | 2.8 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.5 | 3.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.5 | 4.9 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.5 | 26.3 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.5 | 9.9 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.5 | 10.1 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.5 | 4.9 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.5 | 4.7 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.5 | 1.4 | GO:1901804 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.5 | 2.3 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.5 | 12.9 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.4 | 3.6 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.4 | 17.3 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.4 | 3.9 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.4 | 5.7 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.4 | 13.8 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.4 | 52.9 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.4 | 14.1 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.4 | 8.0 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.4 | 2.8 | GO:0001705 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) |
| 0.4 | 4.8 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.4 | 39.0 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.4 | 5.8 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.4 | 13.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.4 | 9.9 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.4 | 1.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.4 | 21.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.4 | 20.6 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.4 | 13.4 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.4 | 7.0 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.4 | 5.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.4 | 13.9 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.4 | 28.9 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.4 | 2.5 | GO:0071569 | protein ufmylation(GO:0071569) protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.4 | 7.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.4 | 6.1 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.4 | 14.0 | GO:0006582 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.4 | 22.9 | GO:0006901 | vesicle coating(GO:0006901) vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.3 | 4.2 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.3 | 8.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.3 | 14.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.3 | 6.0 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 0.3 | 1.3 | GO:0033306 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.3 | 0.9 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.3 | 24.5 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.3 | 11.2 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.3 | 1.8 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.3 | 18.6 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.3 | 1.7 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.3 | 7.5 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.3 | 13.7 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.3 | 1.8 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.3 | 9.6 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.2 | 8.6 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.2 | 1.4 | GO:0060356 | leucine import(GO:0060356) |
| 0.2 | 6.9 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.2 | 16.6 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.2 | 2.9 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.2 | 2.1 | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902237) |
| 0.2 | 5.0 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.2 | 5.1 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.2 | 5.7 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.2 | 3.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 3.0 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) |
| 0.2 | 16.0 | GO:2000177 | regulation of neural precursor cell proliferation(GO:2000177) |
| 0.2 | 13.8 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.2 | 79.5 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.2 | 0.7 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.2 | 4.3 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.2 | 0.5 | GO:0050720 | interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.2 | 1.6 | GO:0090656 | t-circle formation(GO:0090656) |
| 0.2 | 0.9 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.2 | 3.2 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 7.7 | GO:0035065 | regulation of histone acetylation(GO:0035065) |
| 0.1 | 11.0 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.1 | 4.0 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.1 | 2.8 | GO:0097502 | mannosylation(GO:0097502) |
| 0.1 | 15.0 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.1 | 3.6 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.1 | 4.4 | GO:0045661 | regulation of myoblast differentiation(GO:0045661) |
| 0.1 | 15.2 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 | 7.3 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 1.5 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 0.5 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 3.4 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 4.4 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.1 | 1.3 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 2.1 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.1 | 0.5 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.1 | 3.3 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 1.4 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.5 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.1 | 2.0 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.1 | 13.3 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 1.2 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 6.0 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.7 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.4 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.7 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.0 | 1.8 | GO:0019827 | stem cell population maintenance(GO:0019827) |
| 0.0 | 3.8 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 1.2 | GO:0050918 | positive chemotaxis(GO:0050918) |
| 0.0 | 0.2 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.8 | GO:0006968 | cellular defense response(GO:0006968) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.1 | 56.4 | GO:1990423 | RZZ complex(GO:1990423) |
| 5.6 | 16.8 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 5.5 | 61.0 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 5.4 | 27.1 | GO:0033503 | HULC complex(GO:0033503) |
| 5.2 | 26.2 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 5.2 | 51.8 | GO:0097255 | R2TP complex(GO:0097255) |
| 5.2 | 15.5 | GO:0055087 | Ski complex(GO:0055087) |
| 3.8 | 23.0 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 3.8 | 19.0 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 3.5 | 21.2 | GO:1990357 | terminal web(GO:1990357) |
| 3.2 | 48.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 3.2 | 9.7 | GO:1990913 | sperm head plasma membrane(GO:1990913) ooplasm(GO:1990917) |
| 3.0 | 9.0 | GO:0034455 | t-UTP complex(GO:0034455) |
| 2.8 | 36.9 | GO:0005688 | U6 snRNP(GO:0005688) |
| 2.7 | 24.3 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 2.6 | 5.1 | GO:1903349 | omegasome membrane(GO:1903349) |
| 2.5 | 29.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 2.4 | 28.4 | GO:0042555 | MCM complex(GO:0042555) |
| 2.3 | 11.7 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 2.3 | 15.9 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 2.2 | 13.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 2.1 | 12.7 | GO:0061617 | MICOS complex(GO:0061617) |
| 2.0 | 10.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 1.8 | 25.7 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 1.8 | 21.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 1.7 | 27.9 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 1.6 | 8.0 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 1.5 | 9.0 | GO:0070545 | PeBoW complex(GO:0070545) |
| 1.4 | 12.8 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 1.4 | 7.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 1.4 | 9.9 | GO:0005683 | U7 snRNP(GO:0005683) |
| 1.3 | 1.3 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 1.3 | 3.9 | GO:0044393 | microspike(GO:0044393) |
| 1.3 | 10.2 | GO:0070187 | telosome(GO:0070187) |
| 1.2 | 8.6 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 1.2 | 33.8 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 1.2 | 22.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 1.1 | 10.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 1.1 | 23.5 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 1.0 | 15.3 | GO:0000346 | transcription export complex(GO:0000346) |
| 1.0 | 4.0 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 1.0 | 20.9 | GO:0097342 | ripoptosome(GO:0097342) |
| 1.0 | 11.8 | GO:0044292 | dendrite terminus(GO:0044292) |
| 1.0 | 58.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 1.0 | 8.6 | GO:0001739 | sex chromatin(GO:0001739) |
| 1.0 | 12.5 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 1.0 | 5.7 | GO:0005816 | spindle pole body(GO:0005816) |
| 0.9 | 12.0 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.9 | 6.4 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.9 | 28.1 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.9 | 21.0 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.9 | 60.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.8 | 7.4 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.8 | 3.3 | GO:0071920 | cleavage body(GO:0071920) |
| 0.8 | 6.5 | GO:0051286 | cell tip(GO:0051286) |
| 0.7 | 5.5 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.7 | 13.9 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.7 | 4.6 | GO:0016589 | NURF complex(GO:0016589) |
| 0.6 | 11.0 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.6 | 2.5 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.6 | 5.0 | GO:0034709 | methylosome(GO:0034709) |
| 0.6 | 9.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.6 | 1.7 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.6 | 15.8 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.5 | 7.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.5 | 4.4 | GO:0030891 | VCB complex(GO:0030891) |
| 0.5 | 8.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.5 | 27.9 | GO:0002102 | podosome(GO:0002102) |
| 0.5 | 2.4 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.5 | 3.9 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.5 | 12.9 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.5 | 21.1 | GO:0005844 | polysome(GO:0005844) |
| 0.5 | 5.0 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.4 | 5.7 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.4 | 23.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.4 | 7.3 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.4 | 10.7 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.4 | 23.5 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.3 | 16.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.3 | 24.1 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.3 | 2.9 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.3 | 1.2 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.3 | 0.9 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.3 | 39.2 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.3 | 17.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 19.9 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.2 | 24.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 35.0 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.2 | 0.6 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.2 | 20.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 4.7 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.2 | 7.9 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.2 | 2.0 | GO:0044666 | MLL3/4 complex(GO:0044666) Set1C/COMPASS complex(GO:0048188) |
| 0.2 | 4.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.2 | 2.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.2 | 1.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 24.9 | GO:0030496 | midbody(GO:0030496) |
| 0.2 | 0.5 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.2 | 0.9 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.2 | 7.4 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 15.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 5.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 6.2 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 4.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 3.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.5 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.1 | 1.4 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 45.4 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 4.5 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 11.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 35.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 11.7 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 6.5 | GO:0031970 | organelle envelope lumen(GO:0031970) |
| 0.1 | 15.9 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.1 | 8.6 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 4.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 2.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 4.5 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 3.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 3.4 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 35.4 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 19.1 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 5.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 8.9 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.4 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 5.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.1 | GO:0005681 | spliceosomal complex(GO:0005681) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.1 | 30.3 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 7.8 | 23.5 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 7.7 | 23.0 | GO:0003896 | DNA primase activity(GO:0003896) |
| 7.1 | 21.4 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 6.1 | 24.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 6.0 | 18.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 5.9 | 17.7 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 5.9 | 17.6 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 4.7 | 23.6 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 4.7 | 14.0 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 4.2 | 12.7 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 4.1 | 12.3 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 4.1 | 41.0 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 4.1 | 16.3 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 4.0 | 15.9 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 3.7 | 11.1 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 3.7 | 11.0 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 3.6 | 14.4 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 3.4 | 10.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 3.3 | 13.4 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 3.3 | 16.6 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 3.2 | 12.9 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 3.0 | 23.8 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 2.8 | 11.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 2.8 | 13.8 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 2.7 | 10.8 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 2.6 | 37.0 | GO:0046790 | virion binding(GO:0046790) |
| 2.6 | 18.4 | GO:0015217 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) ATP transmembrane transporter activity(GO:0005347) purine nucleotide transmembrane transporter activity(GO:0015216) ADP transmembrane transporter activity(GO:0015217) |
| 2.5 | 20.3 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 2.5 | 20.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 2.5 | 7.5 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 2.5 | 9.9 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 2.3 | 11.7 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 2.3 | 6.9 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 2.0 | 10.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 2.0 | 8.1 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 1.9 | 15.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 1.7 | 10.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 1.7 | 47.1 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 1.6 | 9.6 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 1.6 | 29.5 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 1.5 | 13.1 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 1.4 | 55.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 1.4 | 19.5 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 1.3 | 8.8 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 1.2 | 4.9 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 1.2 | 7.4 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 1.2 | 36.8 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 1.1 | 4.6 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 1.1 | 12.5 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 1.1 | 9.8 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 1.1 | 9.8 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 1.1 | 53.9 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 1.0 | 26.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 1.0 | 4.9 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 1.0 | 20.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.9 | 25.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.9 | 8.3 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.9 | 27.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.9 | 19.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.8 | 6.6 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.8 | 4.8 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.8 | 16.9 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.8 | 2.3 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.7 | 20.8 | GO:0001848 | complement binding(GO:0001848) |
| 0.7 | 3.5 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.7 | 48.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.7 | 2.8 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.7 | 8.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.6 | 3.2 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.6 | 8.6 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.6 | 2.4 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.6 | 4.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.6 | 11.7 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.6 | 1.7 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.6 | 24.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.6 | 17.0 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.6 | 5.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.5 | 30.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.5 | 8.7 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.5 | 16.1 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.5 | 13.9 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.5 | 13.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.5 | 13.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.5 | 39.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.5 | 30.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.5 | 7.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.5 | 2.4 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.5 | 6.0 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.5 | 1.4 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.5 | 7.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.4 | 10.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.4 | 13.7 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.4 | 5.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.4 | 1.6 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.4 | 21.8 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.4 | 13.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.4 | 2.3 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.4 | 12.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.4 | 11.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.4 | 2.5 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.4 | 1.4 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.3 | 5.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.3 | 3.6 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.3 | 16.0 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.3 | 12.4 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.3 | 7.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.3 | 1.2 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.3 | 62.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.3 | 7.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.3 | 7.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.3 | 10.1 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.3 | 3.6 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.3 | 8.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.3 | 4.4 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.3 | 1.4 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.3 | 5.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.3 | 14.7 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.2 | 6.7 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 3.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.2 | 12.2 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.2 | 0.7 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.2 | 6.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.2 | 0.6 | GO:0000406 | double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.2 | 11.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.2 | 1.4 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.2 | 6.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 5.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.2 | 3.0 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.2 | 2.8 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 9.0 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.2 | 15.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 0.9 | GO:0089720 | caspase binding(GO:0089720) |
| 0.2 | 9.9 | GO:0042379 | chemokine receptor binding(GO:0042379) |
| 0.2 | 3.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.2 | 2.1 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.2 | 22.9 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.2 | 3.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 10.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.2 | 5.9 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 3.0 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 18.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 3.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.2 | 29.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 4.0 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.2 | 3.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 11.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 3.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 11.4 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 2.0 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 1.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 1.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 18.3 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.1 | 4.1 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 3.7 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 11.4 | GO:0016597 | amino acid binding(GO:0016597) |
| 0.1 | 4.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 7.1 | GO:0019783 | ubiquitin-like protein-specific protease activity(GO:0019783) |
| 0.1 | 4.5 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) ATP-dependent helicase activity(GO:0008026) RNA-dependent ATPase activity(GO:0008186) purine NTP-dependent helicase activity(GO:0070035) |
| 0.1 | 0.7 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 6.3 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 0.4 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.1 | 1.2 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 4.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 16.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 3.3 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 6.5 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 4.2 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 1.4 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 2.7 | GO:0016829 | lyase activity(GO:0016829) |
| 0.0 | 3.7 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 3.5 | GO:0016887 | ATPase activity(GO:0016887) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 61.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 1.5 | 21.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 1.4 | 84.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.8 | 8.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.8 | 24.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.7 | 43.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.7 | 18.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.6 | 18.7 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.6 | 18.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.6 | 20.2 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.5 | 39.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.5 | 26.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.4 | 10.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.4 | 13.9 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.4 | 31.0 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.4 | 15.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.4 | 13.1 | PID MYC PATHWAY | C-MYC pathway |
| 0.4 | 12.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.4 | 12.5 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.2 | 16.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.2 | 10.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 10.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.2 | 17.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.2 | 4.7 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 10.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.2 | 27.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.2 | 4.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.2 | 8.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 11.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 11.9 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.1 | 9.7 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 4.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 3.2 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 3.3 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 9.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 3.0 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 2.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 10.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 3.0 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.9 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 44.4 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 2.7 | 38.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 2.5 | 61.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 2.0 | 30.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 2.0 | 56.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 1.9 | 13.1 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 1.8 | 3.6 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 1.5 | 113.0 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 1.4 | 54.5 | REACTOME G2 M CHECKPOINTS | Genes involved in G2/M Checkpoints |
| 1.3 | 10.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 1.2 | 19.9 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 1.2 | 29.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 1.1 | 45.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.9 | 18.8 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.9 | 21.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.8 | 3.3 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.8 | 24.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.7 | 21.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.7 | 10.9 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.7 | 9.9 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.7 | 20.9 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.7 | 11.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.6 | 18.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.6 | 22.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.6 | 17.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.6 | 5.7 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.6 | 10.0 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.5 | 15.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.5 | 57.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.5 | 43.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.5 | 12.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.5 | 7.5 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.4 | 5.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.4 | 13.0 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.4 | 8.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.4 | 9.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.4 | 8.5 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.4 | 11.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.4 | 17.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.4 | 13.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.3 | 4.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.3 | 5.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.3 | 37.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.3 | 10.0 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.3 | 21.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.3 | 35.0 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.3 | 8.1 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.2 | 5.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 4.7 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.2 | 10.4 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.2 | 8.6 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.2 | 7.9 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.2 | 4.0 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.2 | 14.0 | REACTOME PI METABOLISM | Genes involved in PI Metabolism |
| 0.2 | 3.3 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.2 | 4.9 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 11.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.2 | 7.4 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.2 | 31.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 3.0 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 7.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 5.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 17.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 3.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 1.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 3.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 3.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 4.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.5 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |