Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
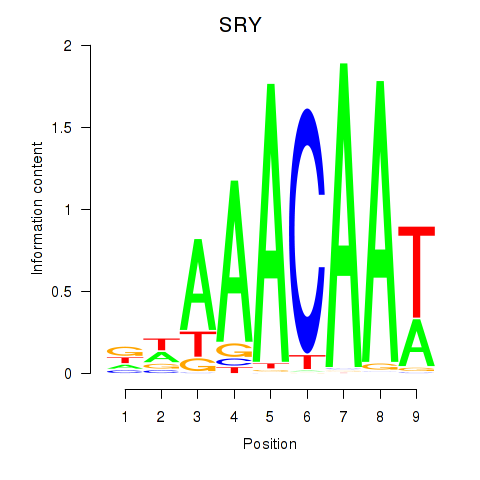
Results for SRY
Z-value: 1.02
Transcription factors associated with SRY
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SRY
|
ENSG00000184895.8 | SRY |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SRY | hg38_v1_chrY_-_2787676_2787687 | 0.35 | 1.0e-07 | Click! |
Activity profile of SRY motif
Sorted Z-values of SRY motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SRY
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_39467672 | 43.94 |
ENST00000436143.6
ENST00000441980.6 ENST00000682069.1 ENST00000311042.10 |
MOBP
|
myelin associated oligodendrocyte basic protein |
| chr3_+_39467598 | 40.44 |
ENST00000428261.5
ENST00000420739.5 ENST00000415443.5 ENST00000447324.5 ENST00000383754.7 |
MOBP
|
myelin associated oligodendrocyte basic protein |
| chr14_-_59630582 | 20.46 |
ENST00000395090.5
|
RTN1
|
reticulon 1 |
| chr10_+_11164961 | 20.15 |
ENST00000399850.7
ENST00000417956.6 |
CELF2
|
CUGBP Elav-like family member 2 |
| chr17_-_64390852 | 17.95 |
ENST00000563924.6
|
PECAM1
|
platelet and endothelial cell adhesion molecule 1 |
| chr9_+_136980211 | 17.83 |
ENST00000444903.2
|
PTGDS
|
prostaglandin D2 synthase |
| chr10_+_69088096 | 14.68 |
ENST00000242465.4
|
SRGN
|
serglycin |
| chr6_-_32589833 | 13.25 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr5_-_147056010 | 12.82 |
ENST00000394414.5
|
PPP2R2B
|
protein phosphatase 2 regulatory subunit Bbeta |
| chr1_-_156248084 | 12.58 |
ENST00000652405.1
ENST00000335852.6 ENST00000540423.5 ENST00000612424.4 ENST00000613336.4 ENST00000623241.3 |
PAQR6
|
progestin and adipoQ receptor family member 6 |
| chr6_-_152168291 | 12.52 |
ENST00000354674.5
|
SYNE1
|
spectrin repeat containing nuclear envelope protein 1 |
| chr5_-_147055968 | 12.46 |
ENST00000336640.10
|
PPP2R2B
|
protein phosphatase 2 regulatory subunit Bbeta |
| chr6_-_152168349 | 12.36 |
ENST00000539504.5
|
SYNE1
|
spectrin repeat containing nuclear envelope protein 1 |
| chr1_-_156248013 | 12.28 |
ENST00000368270.2
|
PAQR6
|
progestin and adipoQ receptor family member 6 |
| chr18_-_55589795 | 12.25 |
ENST00000568740.5
ENST00000629387.2 |
TCF4
|
transcription factor 4 |
| chr1_-_156248038 | 12.07 |
ENST00000470198.5
ENST00000292291.10 ENST00000356983.7 |
PAQR6
|
progestin and adipoQ receptor family member 6 |
| chr11_-_134412234 | 11.89 |
ENST00000312527.9
|
B3GAT1
|
beta-1,3-glucuronyltransferase 1 |
| chr18_-_55635948 | 11.28 |
ENST00000565124.4
ENST00000398339.5 |
TCF4
|
transcription factor 4 |
| chr18_-_55589836 | 11.28 |
ENST00000537578.5
ENST00000564403.6 |
TCF4
|
transcription factor 4 |
| chr18_-_55321986 | 11.28 |
ENST00000570287.6
|
TCF4
|
transcription factor 4 |
| chr18_-_55589770 | 11.14 |
ENST00000565018.6
ENST00000636400.2 |
TCF4
|
transcription factor 4 |
| chr5_-_147081428 | 11.12 |
ENST00000394413.7
|
PPP2R2B
|
protein phosphatase 2 regulatory subunit Bbeta |
| chr10_+_11165475 | 11.06 |
ENST00000609692.5
ENST00000354897.3 |
CELF2
|
CUGBP Elav-like family member 2 |
| chr2_-_71227055 | 10.86 |
ENST00000244221.9
|
PAIP2B
|
poly(A) binding protein interacting protein 2B |
| chr3_+_189789672 | 10.57 |
ENST00000434928.5
|
TP63
|
tumor protein p63 |
| chr11_+_114060204 | 10.56 |
ENST00000683318.1
|
ZBTB16
|
zinc finger and BTB domain containing 16 |
| chr10_+_110207587 | 10.37 |
ENST00000332674.9
ENST00000453116.5 |
MXI1
|
MAX interactor 1, dimerization protein |
| chr21_-_31558977 | 10.36 |
ENST00000286827.7
ENST00000541036.5 |
TIAM1
|
TIAM Rac1 associated GEF 1 |
| chr3_+_115623502 | 10.20 |
ENST00000305124.11
ENST00000393780.3 |
GAP43
|
growth associated protein 43 |
| chr5_-_131796921 | 9.77 |
ENST00000307968.11
ENST00000307954.12 |
FNIP1
|
folliculin interacting protein 1 |
| chr11_+_131911396 | 9.56 |
ENST00000425719.6
ENST00000374784.5 |
NTM
|
neurotrimin |
| chr22_+_19723525 | 9.49 |
ENST00000366425.4
|
GP1BB
|
glycoprotein Ib platelet subunit beta |
| chr17_-_17972374 | 9.35 |
ENST00000318094.14
ENST00000540946.5 ENST00000379504.8 ENST00000542206.5 ENST00000395739.8 ENST00000581396.5 ENST00000535933.5 ENST00000579586.1 |
TOM1L2
|
target of myb1 like 2 membrane trafficking protein |
| chr16_-_1782526 | 9.19 |
ENST00000566339.6
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3 |
| chr19_-_45768627 | 9.08 |
ENST00000560160.1
|
SIX5
|
SIX homeobox 5 |
| chr17_+_17972813 | 8.93 |
ENST00000582416.5
ENST00000313838.12 ENST00000399187.6 ENST00000581264.5 ENST00000479684.2 ENST00000584166.5 ENST00000585108.5 ENST00000399182.5 ENST00000579977.1 |
DRC3
|
dynein regulatory complex subunit 3 |
| chr5_-_11589019 | 8.77 |
ENST00000511377.5
|
CTNND2
|
catenin delta 2 |
| chr16_+_6019585 | 8.59 |
ENST00000547372.5
|
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr5_-_131797030 | 8.52 |
ENST00000615660.4
|
FNIP1
|
folliculin interacting protein 1 |
| chr11_+_112961247 | 8.38 |
ENST00000621518.4
ENST00000618266.4 ENST00000615112.4 ENST00000615285.4 ENST00000619839.4 ENST00000401611.6 ENST00000621128.4 |
NCAM1
|
neural cell adhesion molecule 1 |
| chr18_-_55322215 | 8.30 |
ENST00000457482.7
|
TCF4
|
transcription factor 4 |
| chr10_+_110225955 | 8.29 |
ENST00000239007.11
|
MXI1
|
MAX interactor 1, dimerization protein |
| chr2_-_2331336 | 7.96 |
ENST00000648933.1
ENST00000644820.1 |
MYT1L
|
myelin transcription factor 1 like |
| chr5_-_147081462 | 7.84 |
ENST00000508267.5
ENST00000504198.5 |
PPP2R2B
|
protein phosphatase 2 regulatory subunit Bbeta |
| chr17_-_39607876 | 7.83 |
ENST00000302584.5
|
NEUROD2
|
neuronal differentiation 2 |
| chrX_+_108045050 | 7.55 |
ENST00000458383.1
ENST00000217957.10 |
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr1_+_50105666 | 7.54 |
ENST00000651347.1
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr2_-_156342348 | 7.49 |
ENST00000409572.5
|
NR4A2
|
nuclear receptor subfamily 4 group A member 2 |
| chr18_-_55321640 | 7.27 |
ENST00000637169.2
|
TCF4
|
transcription factor 4 |
| chrX_-_107716401 | 7.16 |
ENST00000486554.1
ENST00000372390.8 |
TSC22D3
|
TSC22 domain family member 3 |
| chr1_+_84164370 | 7.01 |
ENST00000446538.5
ENST00000610703.4 ENST00000370682.7 ENST00000394838.6 ENST00000432111.5 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chrX_+_108044967 | 6.96 |
ENST00000415430.7
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr4_+_164754116 | 6.96 |
ENST00000507311.1
|
SMIM31
|
small integral membrane protein 31 |
| chr11_+_128694052 | 6.91 |
ENST00000527786.7
ENST00000534087.3 |
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr18_-_55302613 | 6.88 |
ENST00000561831.7
|
TCF4
|
transcription factor 4 |
| chr1_+_50106265 | 6.82 |
ENST00000357083.8
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr18_+_74534493 | 6.79 |
ENST00000358821.8
|
CNDP1
|
carnosine dipeptidase 1 |
| chr14_+_36657560 | 6.77 |
ENST00000402703.6
|
PAX9
|
paired box 9 |
| chr2_-_86337617 | 6.65 |
ENST00000538924.7
ENST00000535845.6 |
REEP1
|
receptor accessory protein 1 |
| chr1_+_50108856 | 6.63 |
ENST00000650764.1
ENST00000494555.2 ENST00000371824.7 ENST00000371823.8 ENST00000652693.1 |
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr10_-_60389833 | 6.62 |
ENST00000280772.7
|
ANK3
|
ankyrin 3 |
| chr2_-_2331225 | 6.61 |
ENST00000648627.1
ENST00000649663.1 ENST00000650560.1 ENST00000428368.7 ENST00000648316.1 ENST00000648665.1 ENST00000649313.1 ENST00000399161.7 ENST00000647738.2 |
MYT1L
|
myelin transcription factor 1 like |
| chr11_+_126355634 | 6.53 |
ENST00000227495.10
ENST00000676545.1 ENST00000678865.1 ENST00000444328.7 ENST00000677503.1 |
ST3GAL4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr18_+_74534594 | 6.47 |
ENST00000582365.1
|
CNDP1
|
carnosine dipeptidase 1 |
| chr2_-_207165923 | 6.26 |
ENST00000309446.11
|
KLF7
|
Kruppel like factor 7 |
| chr2_-_86337654 | 6.19 |
ENST00000165698.9
|
REEP1
|
receptor accessory protein 1 |
| chr21_-_31160904 | 6.08 |
ENST00000636887.1
|
TIAM1
|
TIAM Rac1 associated GEF 1 |
| chr5_-_131796965 | 6.01 |
ENST00000514667.1
ENST00000511848.1 ENST00000510461.6 |
ENSG00000273217.1
FNIP1
|
novel protein folliculin interacting protein 1 |
| chr8_+_84183262 | 5.91 |
ENST00000522455.5
ENST00000521695.5 ENST00000521268.6 |
RALYL
|
RALY RNA binding protein like |
| chr1_+_84164962 | 5.90 |
ENST00000614872.4
ENST00000394839.6 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr16_+_6019016 | 5.76 |
ENST00000550418.6
|
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr8_+_84183534 | 5.75 |
ENST00000518566.5
|
RALYL
|
RALY RNA binding protein like |
| chr11_+_128693887 | 5.67 |
ENST00000281428.12
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr14_+_91114026 | 5.61 |
ENST00000521081.5
ENST00000520328.5 ENST00000524232.5 ENST00000522170.5 ENST00000256324.15 ENST00000519950.5 ENST00000523879.5 ENST00000521077.6 ENST00000518665.6 |
DGLUCY
|
D-glutamate cyclase |
| chrX_+_85244075 | 5.53 |
ENST00000276123.7
|
ZNF711
|
zinc finger protein 711 |
| chr16_-_68448491 | 5.53 |
ENST00000561749.1
ENST00000219334.10 |
SMPD3
|
sphingomyelin phosphodiesterase 3 |
| chr11_+_112961402 | 5.46 |
ENST00000613217.4
ENST00000316851.12 ENST00000620046.4 ENST00000531044.5 ENST00000529356.5 |
NCAM1
|
neural cell adhesion molecule 1 |
| chr1_-_177164673 | 5.45 |
ENST00000424564.2
ENST00000361833.7 |
ASTN1
|
astrotactin 1 |
| chr16_+_2537997 | 5.32 |
ENST00000441549.7
ENST00000268673.11 ENST00000342085.9 ENST00000389224.7 |
PDPK1
|
3-phosphoinositide dependent protein kinase 1 |
| chr16_-_15643024 | 5.32 |
ENST00000540441.6
|
MARF1
|
meiosis regulator and mRNA stability factor 1 |
| chr14_+_91114431 | 5.23 |
ENST00000428926.6
ENST00000517362.5 |
DGLUCY
|
D-glutamate cyclase |
| chr20_-_3781440 | 5.17 |
ENST00000379756.3
|
SPEF1
|
sperm flagellar 1 |
| chr16_+_6019071 | 5.17 |
ENST00000547605.5
ENST00000553186.5 |
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr9_-_23826299 | 5.13 |
ENST00000380117.5
|
ELAVL2
|
ELAV like RNA binding protein 2 |
| chr15_-_34318761 | 5.11 |
ENST00000290209.9
|
SLC12A6
|
solute carrier family 12 member 6 |
| chr14_+_91114388 | 5.10 |
ENST00000519019.5
ENST00000523816.5 ENST00000517518.5 |
DGLUCY
|
D-glutamate cyclase |
| chr11_+_112961480 | 5.10 |
ENST00000621850.4
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr13_-_40666600 | 5.04 |
ENST00000379561.6
|
FOXO1
|
forkhead box O1 |
| chr1_-_151146643 | 5.03 |
ENST00000613223.1
|
SEMA6C
|
semaphorin 6C |
| chr2_-_2326378 | 5.02 |
ENST00000647618.1
|
MYT1L
|
myelin transcription factor 1 like |
| chr17_+_79778135 | 4.91 |
ENST00000310942.9
ENST00000269399.5 |
CBX2
|
chromobox 2 |
| chr14_+_91114364 | 4.85 |
ENST00000518868.5
|
DGLUCY
|
D-glutamate cyclase |
| chr10_-_60572599 | 4.82 |
ENST00000503366.5
|
ANK3
|
ankyrin 3 |
| chr5_+_140868945 | 4.78 |
ENST00000398640.7
|
PCDHA11
|
protocadherin alpha 11 |
| chr4_-_173530219 | 4.69 |
ENST00000359562.4
|
HAND2
|
heart and neural crest derivatives expressed 2 |
| chrX_+_85244032 | 4.67 |
ENST00000373165.7
|
ZNF711
|
zinc finger protein 711 |
| chr13_-_46142834 | 4.62 |
ENST00000674665.1
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr5_+_141364231 | 4.62 |
ENST00000611914.1
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chr5_-_150086511 | 4.55 |
ENST00000675795.1
|
CSF1R
|
colony stimulating factor 1 receptor |
| chr1_+_66332004 | 4.52 |
ENST00000371045.9
ENST00000531025.5 ENST00000526197.5 |
PDE4B
|
phosphodiesterase 4B |
| chr15_-_85794902 | 4.51 |
ENST00000337975.6
|
KLHL25
|
kelch like family member 25 |
| chr14_+_91114667 | 4.49 |
ENST00000523894.5
ENST00000522322.5 ENST00000523771.5 |
DGLUCY
|
D-glutamate cyclase |
| chr5_+_66958870 | 4.48 |
ENST00000405643.5
ENST00000407621.1 ENST00000432426.5 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr8_-_133297092 | 4.35 |
ENST00000522890.5
ENST00000675983.1 ENST00000518176.5 ENST00000323851.13 ENST00000522476.5 ENST00000518066.5 ENST00000521544.5 ENST00000674605.1 ENST00000518480.5 ENST00000523892.5 |
NDRG1
|
N-myc downstream regulated 1 |
| chr1_+_147902789 | 4.26 |
ENST00000369235.2
|
GJA8
|
gap junction protein alpha 8 |
| chr1_-_151146611 | 4.18 |
ENST00000341697.7
ENST00000368914.8 |
SEMA6C
|
semaphorin 6C |
| chr12_+_94148553 | 4.00 |
ENST00000258526.9
|
PLXNC1
|
plexin C1 |
| chr2_-_165203870 | 3.98 |
ENST00000639244.1
ENST00000409101.7 ENST00000668657.1 |
SCN3A
|
sodium voltage-gated channel alpha subunit 3 |
| chr7_+_29479712 | 3.98 |
ENST00000412711.6
|
CHN2
|
chimerin 2 |
| chr11_-_82997477 | 3.94 |
ENST00000534301.5
|
RAB30
|
RAB30, member RAS oncogene family |
| chr10_+_91162958 | 3.92 |
ENST00000614189.4
|
PCGF5
|
polycomb group ring finger 5 |
| chr10_-_60141004 | 3.91 |
ENST00000355288.6
|
ANK3
|
ankyrin 3 |
| chr7_+_29480077 | 3.86 |
ENST00000439711.6
ENST00000421775.6 ENST00000424025.4 ENST00000409041.7 |
CHN2
|
chimerin 2 |
| chr14_-_22819721 | 3.81 |
ENST00000554517.5
ENST00000285850.11 ENST00000397529.6 ENST00000555702.5 |
SLC7A7
|
solute carrier family 7 member 7 |
| chr12_+_11649666 | 3.71 |
ENST00000396373.9
|
ETV6
|
ETS variant transcription factor 6 |
| chr2_-_165204042 | 3.70 |
ENST00000283254.12
ENST00000453007.1 |
SCN3A
|
sodium voltage-gated channel alpha subunit 3 |
| chr2_+_86720282 | 3.70 |
ENST00000283632.5
|
RMND5A
|
required for meiotic nuclear division 5 homolog A |
| chr2_-_144516397 | 3.66 |
ENST00000638128.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr9_-_23826231 | 3.66 |
ENST00000397312.7
|
ELAVL2
|
ELAV like RNA binding protein 2 |
| chr18_-_55351977 | 3.64 |
ENST00000643689.1
|
TCF4
|
transcription factor 4 |
| chr2_-_144516154 | 3.64 |
ENST00000637304.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr7_+_80638633 | 3.58 |
ENST00000447544.7
ENST00000482059.6 |
CD36
|
CD36 molecule |
| chrX_-_111412162 | 3.57 |
ENST00000637570.1
ENST00000356220.8 ENST00000636035.2 ENST00000637453.1 ENST00000635795.1 |
DCX
|
doublecortin |
| chr8_+_24294044 | 3.51 |
ENST00000265769.9
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr8_-_71547626 | 3.50 |
ENST00000647540.1
ENST00000644229.1 |
EYA1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr19_-_45769204 | 3.32 |
ENST00000317578.7
|
SIX5
|
SIX homeobox 5 |
| chr7_-_78771265 | 3.30 |
ENST00000630991.2
ENST00000629359.2 |
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr3_-_28348805 | 3.22 |
ENST00000457172.5
ENST00000479665.6 |
AZI2
|
5-azacytidine induced 2 |
| chr9_+_2110354 | 3.21 |
ENST00000634772.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr3_+_124384513 | 3.15 |
ENST00000682540.1
ENST00000522553.6 ENST00000682695.1 ENST00000682674.1 ENST00000684382.1 |
KALRN
|
kalirin RhoGEF kinase |
| chr14_-_89417148 | 3.13 |
ENST00000557258.6
|
FOXN3
|
forkhead box N3 |
| chr2_-_219178103 | 3.09 |
ENST00000409789.5
|
CNPPD1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr1_+_43389889 | 3.08 |
ENST00000562955.2
ENST00000634258.3 |
SZT2
|
SZT2 subunit of KICSTOR complex |
| chr1_-_56579555 | 3.08 |
ENST00000371250.4
|
PLPP3
|
phospholipid phosphatase 3 |
| chr5_-_24644968 | 3.05 |
ENST00000264463.8
|
CDH10
|
cadherin 10 |
| chr3_-_62373015 | 3.02 |
ENST00000475839.1
|
FEZF2
|
FEZ family zinc finger 2 |
| chr1_-_217137748 | 2.98 |
ENST00000493603.5
ENST00000366940.6 |
ESRRG
|
estrogen related receptor gamma |
| chr7_+_144070313 | 2.98 |
ENST00000641441.1
|
OR2A25
|
olfactory receptor family 2 subfamily A member 25 |
| chr10_+_97319250 | 2.92 |
ENST00000371021.5
|
FRAT1
|
FRAT regulator of WNT signaling pathway 1 |
| chr3_-_33659441 | 2.91 |
ENST00000650653.1
ENST00000480013.6 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr8_-_144462848 | 2.89 |
ENST00000530374.6
|
CYHR1
|
cysteine and histidine rich 1 |
| chr17_-_58328756 | 2.87 |
ENST00000268893.10
ENST00000343736.9 |
TSPOAP1
|
TSPO associated protein 1 |
| chr8_-_71361860 | 2.85 |
ENST00000303824.11
ENST00000645451.1 |
EYA1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr18_+_11851404 | 2.82 |
ENST00000526991.3
|
CHMP1B
|
charged multivesicular body protein 1B |
| chr14_+_74348440 | 2.79 |
ENST00000256362.5
|
VRTN
|
vertebrae development associated |
| chr16_+_28846674 | 2.79 |
ENST00000322610.12
|
SH2B1
|
SH2B adaptor protein 1 |
| chr6_-_162727748 | 2.73 |
ENST00000366892.5
ENST00000366898.6 ENST00000366897.5 ENST00000366896.5 ENST00000674250.1 |
PRKN
|
parkin RBR E3 ubiquitin protein ligase |
| chr5_-_147782681 | 2.71 |
ENST00000616793.5
ENST00000333010.6 ENST00000265272.9 |
JAKMIP2
|
janus kinase and microtubule interacting protein 2 |
| chr11_+_67240047 | 2.70 |
ENST00000530342.2
|
KDM2A
|
lysine demethylase 2A |
| chr12_-_10098977 | 2.68 |
ENST00000315330.8
ENST00000457018.6 |
CLEC1A
|
C-type lectin domain family 1 member A |
| chr1_+_25338294 | 2.67 |
ENST00000374358.5
|
TMEM50A
|
transmembrane protein 50A |
| chr5_+_141364153 | 2.55 |
ENST00000518069.2
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chr9_-_127980976 | 2.52 |
ENST00000373095.6
|
FAM102A
|
family with sequence similarity 102 member A |
| chr2_-_159616442 | 2.47 |
ENST00000541068.6
ENST00000392783.7 ENST00000392782.5 |
BAZ2B
|
bromodomain adjacent to zinc finger domain 2B |
| chr7_+_80638510 | 2.47 |
ENST00000433696.6
ENST00000538969.5 ENST00000544133.5 |
CD36
|
CD36 molecule |
| chr15_+_90872862 | 2.45 |
ENST00000618099.4
|
FURIN
|
furin, paired basic amino acid cleaving enzyme |
| chr13_-_99258366 | 2.43 |
ENST00000397470.5
ENST00000397473.7 |
GPR18
|
G protein-coupled receptor 18 |
| chr4_+_118034480 | 2.43 |
ENST00000296499.6
|
NDST3
|
N-deacetylase and N-sulfotransferase 3 |
| chr21_-_18403754 | 2.42 |
ENST00000284885.8
|
TMPRSS15
|
transmembrane serine protease 15 |
| chr10_+_122374685 | 2.35 |
ENST00000368990.7
ENST00000368989.6 ENST00000463663.6 |
PLEKHA1
|
pleckstrin homology domain containing A1 |
| chr14_+_58427385 | 2.30 |
ENST00000354386.10
ENST00000619416.4 |
KIAA0586
|
KIAA0586 |
| chr8_-_81483226 | 2.29 |
ENST00000256104.5
|
FABP4
|
fatty acid binding protein 4 |
| chr12_+_55019967 | 2.27 |
ENST00000242994.4
|
NEUROD4
|
neuronal differentiation 4 |
| chr12_+_54280842 | 2.27 |
ENST00000678077.1
ENST00000548688.5 |
HNRNPA1
|
heterogeneous nuclear ribonucleoprotein A1 |
| chr17_-_69141878 | 2.23 |
ENST00000590645.1
ENST00000284425.7 |
ABCA6
|
ATP binding cassette subfamily A member 6 |
| chr3_-_33659097 | 2.23 |
ENST00000461133.8
ENST00000496954.2 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr22_+_39901075 | 2.20 |
ENST00000344138.9
|
GRAP2
|
GRB2 related adaptor protein 2 |
| chr14_+_58427425 | 2.18 |
ENST00000619722.5
ENST00000423743.7 |
KIAA0586
|
KIAA0586 |
| chr11_-_119729158 | 2.17 |
ENST00000264025.8
|
NECTIN1
|
nectin cell adhesion molecule 1 |
| chr10_-_97334698 | 2.15 |
ENST00000371019.4
|
FRAT2
|
FRAT regulator of WNT signaling pathway 2 |
| chr21_-_17819386 | 2.14 |
ENST00000400559.7
ENST00000400558.7 |
C21orf91
|
chromosome 21 open reading frame 91 |
| chr11_+_22338333 | 2.14 |
ENST00000263160.4
|
SLC17A6
|
solute carrier family 17 member 6 |
| chr17_+_38705482 | 2.14 |
ENST00000620609.4
|
MLLT6
|
MLLT6, PHD finger containing |
| chrX_+_11760087 | 2.06 |
ENST00000649271.1
|
MSL3
|
MSL complex subunit 3 |
| chr3_+_178536205 | 2.06 |
ENST00000420517.6
|
KCNMB2
|
potassium calcium-activated channel subfamily M regulatory beta subunit 2 |
| chr6_+_31114793 | 2.06 |
ENST00000259881.10
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr12_-_95217373 | 2.05 |
ENST00000549499.1
ENST00000546711.5 ENST00000343958.9 |
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr2_+_219178266 | 2.00 |
ENST00000430297.7
|
RETREG2
|
reticulophagy regulator family member 2 |
| chr6_+_162727941 | 1.91 |
ENST00000366888.6
|
PACRG
|
parkin coregulated |
| chr16_-_4273014 | 1.86 |
ENST00000204517.11
|
TFAP4
|
transcription factor AP-4 |
| chr17_-_42276341 | 1.85 |
ENST00000293328.8
|
STAT5B
|
signal transducer and activator of transcription 5B |
| chr9_-_127847117 | 1.81 |
ENST00000480266.5
|
ENG
|
endoglin |
| chr22_-_31345770 | 1.78 |
ENST00000215919.3
|
PATZ1
|
POZ/BTB and AT hook containing zinc finger 1 |
| chr9_-_37465402 | 1.75 |
ENST00000307750.5
|
ZBTB5
|
zinc finger and BTB domain containing 5 |
| chr11_-_44950151 | 1.75 |
ENST00000533940.5
ENST00000533937.1 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr12_-_7503841 | 1.74 |
ENST00000359156.8
|
CD163
|
CD163 molecule |
| chr21_-_17819343 | 1.71 |
ENST00000284881.9
|
C21orf91
|
chromosome 21 open reading frame 91 |
| chrX_+_11760035 | 1.63 |
ENST00000482871.6
ENST00000648013.1 |
MSL3
|
MSL complex subunit 3 |
| chr1_+_35807974 | 1.53 |
ENST00000373210.4
|
AGO4
|
argonaute RISC component 4 |
| chr4_+_94995919 | 1.51 |
ENST00000509540.6
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr4_+_94974984 | 1.45 |
ENST00000672698.1
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr4_-_67883987 | 1.44 |
ENST00000283916.11
|
TMPRSS11D
|
transmembrane serine protease 11D |
| chr6_+_138773747 | 1.43 |
ENST00000617445.5
|
CCDC28A
|
coiled-coil domain containing 28A |
| chr22_-_49827512 | 1.43 |
ENST00000404760.5
|
BRD1
|
bromodomain containing 1 |
| chr18_+_33578213 | 1.30 |
ENST00000681521.1
ENST00000269197.12 |
ASXL3
|
ASXL transcriptional regulator 3 |
| chr1_-_193186599 | 1.28 |
ENST00000367434.5
|
B3GALT2
|
beta-1,3-galactosyltransferase 2 |
| chr7_+_143288215 | 1.26 |
ENST00000619992.4
ENST00000310447.10 |
CASP2
|
caspase 2 |
| chr11_-_82997394 | 1.24 |
ENST00000525117.5
ENST00000532548.5 |
RAB30
|
RAB30, member RAS oncogene family |
| chr2_+_104853259 | 1.20 |
ENST00000598623.1
ENST00000653688.1 ENST00000662784.1 ENST00000666977.1 ENST00000674056.1 |
ENSG00000269707.2
POU3F3
|
novel transcript, sense overlapping POU3F3 POU class 3 homeobox 3 |
| chr7_+_143959927 | 1.18 |
ENST00000624504.1
|
OR2F1
|
olfactory receptor family 2 subfamily F member 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 84.4 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 4.9 | 14.7 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 4.9 | 24.3 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 4.5 | 17.9 | GO:0072011 | diapedesis(GO:0050904) glomerular endothelium development(GO:0072011) |
| 4.1 | 16.4 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 3.8 | 15.4 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 3.3 | 13.2 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 2.5 | 7.5 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 2.5 | 24.9 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 2.2 | 12.9 | GO:0097338 | response to clozapine(GO:0097338) |
| 2.1 | 8.2 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 2.0 | 7.8 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 1.8 | 10.6 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 1.7 | 40.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 1.4 | 10.1 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 1.3 | 5.0 | GO:1902617 | response to fluoride(GO:1902617) |
| 1.1 | 10.2 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 1.1 | 3.2 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.9 | 4.7 | GO:0061032 | cardiac right ventricle formation(GO:0003219) visceral serous pericardium development(GO:0061032) |
| 0.9 | 7.3 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.9 | 2.7 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.9 | 7.2 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.9 | 5.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.8 | 17.8 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.8 | 2.4 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.7 | 5.1 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.7 | 31.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.7 | 6.3 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.7 | 14.5 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.7 | 6.1 | GO:2000332 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.6 | 3.7 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.6 | 2.4 | GO:0019082 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.6 | 1.8 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.6 | 19.3 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.5 | 12.6 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.5 | 3.8 | GO:1990822 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.5 | 10.1 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.5 | 3.1 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 0.5 | 4.6 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.5 | 5.5 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.4 | 3.1 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.4 | 5.3 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.4 | 3.0 | GO:0043696 | forebrain anterior/posterior pattern specification(GO:0021797) dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.4 | 3.0 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.4 | 1.2 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.4 | 4.3 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.4 | 2.3 | GO:1903936 | cellular response to sodium arsenite(GO:1903936) |
| 0.4 | 4.5 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.4 | 10.9 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.4 | 9.5 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.3 | 1.6 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.3 | 1.8 | GO:0045588 | positive regulation of gamma-delta T cell differentiation(GO:0045588) |
| 0.3 | 1.5 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.3 | 11.9 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.3 | 2.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.3 | 1.5 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.3 | 2.4 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.3 | 0.8 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.3 | 3.1 | GO:0008354 | germ cell migration(GO:0008354) Bergmann glial cell differentiation(GO:0060020) |
| 0.3 | 71.8 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.3 | 9.6 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.2 | 13.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.2 | 4.6 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.2 | 2.2 | GO:0070166 | enamel mineralization(GO:0070166) protein localization to cell junction(GO:1902414) |
| 0.2 | 2.1 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.2 | 2.8 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.2 | 6.8 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.2 | 0.7 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.2 | 6.5 | GO:0097503 | sialylation(GO:0097503) |
| 0.2 | 1.3 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.2 | 3.1 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.2 | 2.7 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.2 | 9.4 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.2 | 5.3 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.2 | 10.9 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.2 | 2.8 | GO:0032196 | transposition(GO:0032196) |
| 0.2 | 4.7 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.2 | 0.5 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.2 | 2.3 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.1 | 3.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 1.3 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 8.8 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.1 | 4.0 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.1 | 18.9 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.1 | 0.2 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 | 4.0 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 0.9 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.1 | 3.4 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.1 | 1.9 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.1 | 2.9 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.1 | 25.1 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.1 | 0.3 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.1 | 0.5 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 0.3 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.1 | 0.5 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 1.4 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 6.2 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 3.6 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 5.4 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 2.1 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 6.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 3.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 19.6 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 0.6 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 2.8 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 3.3 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.2 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.1 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.0 | 0.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 1.2 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 1.7 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 1.4 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 1.8 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.2 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 1.7 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 3.3 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.4 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 12.4 | GO:0032584 | growth cone membrane(GO:0032584) |
| 1.9 | 24.9 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 1.2 | 3.7 | GO:0034657 | GID complex(GO:0034657) |
| 1.0 | 40.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.8 | 4.6 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.7 | 16.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.7 | 13.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.6 | 24.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.6 | 12.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.6 | 5.5 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.6 | 15.4 | GO:0043194 | spectrin-associated cytoskeleton(GO:0014731) axon initial segment(GO:0043194) |
| 0.6 | 1.8 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.6 | 14.7 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.5 | 5.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.5 | 3.7 | GO:0072487 | MSL complex(GO:0072487) |
| 0.5 | 2.7 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.5 | 52.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.4 | 3.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.4 | 10.1 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.4 | 7.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.4 | 76.3 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.3 | 4.9 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.3 | 0.8 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.2 | 3.0 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 1.4 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.2 | 4.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 2.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.2 | 20.9 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.2 | 4.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 4.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 1.5 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.1 | 12.1 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.1 | 6.2 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 1.9 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 27.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 12.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 6.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 13.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 4.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 10.6 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 4.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 26.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 2.1 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 3.2 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 2.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 3.6 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 2.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 84.5 | GO:0016021 | integral component of membrane(GO:0016021) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.0 | 84.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 6.1 | 24.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 4.5 | 17.8 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 4.3 | 12.8 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 4.0 | 83.3 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 3.0 | 11.9 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 1.6 | 6.5 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 1.5 | 4.6 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 1.1 | 9.5 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 1.0 | 2.9 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.9 | 10.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.8 | 7.6 | GO:0043426 | MRF binding(GO:0043426) |
| 0.8 | 24.9 | GO:0005521 | lamin binding(GO:0005521) |
| 0.8 | 12.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.8 | 2.3 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.8 | 6.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.7 | 31.9 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.6 | 8.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.6 | 5.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.6 | 19.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.6 | 13.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.6 | 21.0 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.6 | 2.8 | GO:0004803 | transposase activity(GO:0004803) |
| 0.5 | 10.9 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.5 | 3.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.5 | 40.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.4 | 13.2 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.4 | 2.4 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.4 | 5.1 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.4 | 46.0 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.3 | 3.8 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.3 | 5.3 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.3 | 4.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.3 | 4.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.3 | 5.0 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.2 | 2.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.2 | 15.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 3.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 8.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 4.0 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 7.7 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.2 | 1.8 | GO:0005534 | galactose binding(GO:0005534) |
| 0.2 | 5.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 2.7 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.2 | 1.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.2 | 0.9 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.2 | 7.3 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.2 | 2.4 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.2 | 2.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.2 | 1.2 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.1 | 0.7 | GO:0034648 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 8.5 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 5.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 1.5 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 8.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 14.2 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 9.4 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 8.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 1.3 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.1 | 10.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 3.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 0.3 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 4.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 1.8 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 2.1 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.1 | 0.5 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 2.3 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 1.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 1.2 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 1.3 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 4.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 11.4 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 3.6 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 4.2 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 6.3 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 2.2 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 1.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 13.4 | GO:0004888 | transmembrane signaling receptor activity(GO:0004888) |
| 0.0 | 0.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 19.6 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.5 | 78.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.4 | 22.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.3 | 14.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.3 | 10.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.3 | 17.9 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.2 | 13.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 15.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 19.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 7.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 4.9 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 2.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 3.1 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 15.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 3.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 8.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 4.6 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 3.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 3.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 3.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 5.9 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 3.8 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.1 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 2.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 80.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 1.1 | 17.9 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.7 | 9.5 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.7 | 13.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.4 | 15.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.4 | 15.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 16.9 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.3 | 18.9 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.3 | 11.9 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.2 | 4.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 16.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 6.2 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.2 | 4.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 2.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 4.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 2.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 2.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 3.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 5.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 2.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 3.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 8.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 3.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.3 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.1 | 2.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 8.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 3.6 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 2.1 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.8 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.5 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |