Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
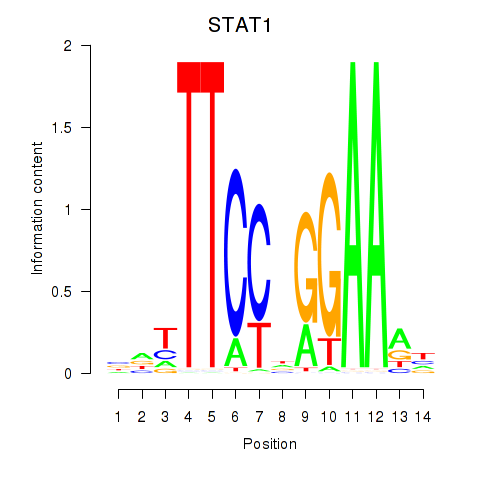
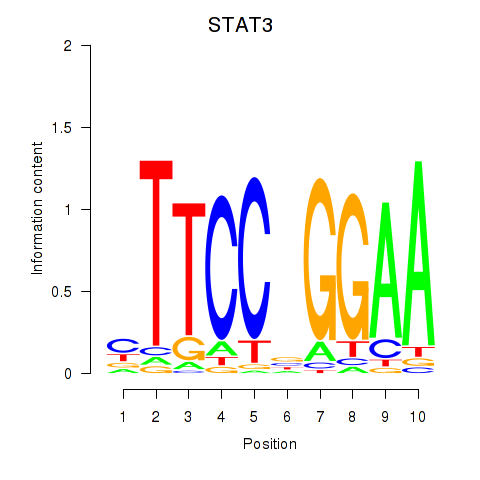
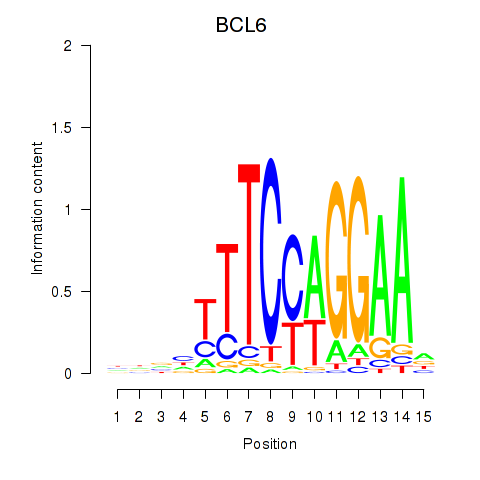
Results for STAT1_STAT3_BCL6
Z-value: 1.17
Transcription factors associated with STAT1_STAT3_BCL6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
STAT1
|
ENSG00000115415.20 | STAT1 |
|
STAT3
|
ENSG00000168610.16 | STAT3 |
|
BCL6
|
ENSG00000113916.18 | BCL6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| STAT1 | hg38_v1_chr2_-_191014137_191014231 | -0.33 | 4.2e-07 | Click! |
| BCL6 | hg38_v1_chr3_-_187736493_187736569 | 0.33 | 4.7e-07 | Click! |
| STAT3 | hg38_v1_chr17_-_42388360_42388463 | 0.05 | 4.9e-01 | Click! |
Activity profile of STAT1_STAT3_BCL6 motif
Sorted Z-values of STAT1_STAT3_BCL6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of STAT1_STAT3_BCL6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.0 | 64.9 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 11.6 | 34.8 | GO:0060739 | mesenchymal-epithelial cell signaling involved in prostate gland development(GO:0060739) |
| 10.3 | 30.8 | GO:0071139 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 9.4 | 28.2 | GO:0014873 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 6.7 | 20.2 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 6.3 | 18.8 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 5.8 | 29.2 | GO:0038123 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 3.1 | 9.3 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 2.5 | 9.8 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 2.4 | 9.8 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 2.1 | 39.8 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 2.0 | 8.2 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 1.9 | 5.7 | GO:0072277 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 1.8 | 5.5 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) response to iron ion starvation(GO:1990641) |
| 1.8 | 19.8 | GO:0002765 | immune response-inhibiting signal transduction(GO:0002765) immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 1.8 | 23.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 1.7 | 26.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 1.7 | 8.7 | GO:0030070 | insulin processing(GO:0030070) |
| 1.7 | 5.2 | GO:0072683 | T cell extravasation(GO:0072683) response to cyclosporin A(GO:1905237) |
| 1.6 | 9.6 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 1.4 | 41.0 | GO:0014850 | response to muscle activity(GO:0014850) |
| 1.4 | 5.6 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 1.4 | 4.1 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) |
| 1.3 | 3.9 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 1.3 | 7.7 | GO:0035604 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) |
| 1.3 | 7.6 | GO:0022614 | T cell antigen processing and presentation(GO:0002457) membrane to membrane docking(GO:0022614) |
| 1.2 | 7.5 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 1.2 | 4.9 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 1.2 | 2.5 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 1.1 | 5.3 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 1.1 | 9.5 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 1.0 | 11.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 1.0 | 1.0 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.9 | 5.3 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.9 | 2.7 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.8 | 5.7 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.8 | 8.6 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.8 | 2.3 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.8 | 3.8 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.7 | 15.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.7 | 3.3 | GO:2000542 | negative regulation of gastrulation(GO:2000542) |
| 0.7 | 4.0 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.7 | 8.5 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.6 | 5.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.6 | 6.3 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.6 | 2.5 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.6 | 2.4 | GO:2000845 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.6 | 1.8 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.6 | 1.8 | GO:1990009 | response to high light intensity(GO:0009644) cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) retinal cell apoptotic process(GO:1990009) |
| 0.6 | 2.3 | GO:0035712 | T-helper 2 cell activation(GO:0035712) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.6 | 2.8 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.5 | 6.0 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.5 | 10.8 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.5 | 1.6 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.5 | 3.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.5 | 4.8 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.5 | 4.8 | GO:0060613 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) fat pad development(GO:0060613) positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) positive regulation of protein monoubiquitination(GO:1902527) |
| 0.5 | 4.6 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.5 | 52.3 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.5 | 3.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.5 | 3.5 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.5 | 10.0 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.5 | 13.1 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.5 | 4.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.5 | 2.8 | GO:1904304 | regulation of gastro-intestinal system smooth muscle contraction(GO:1904304) positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.5 | 2.8 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.5 | 2.7 | GO:0006050 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.5 | 22.2 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.5 | 5.0 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.5 | 4.5 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.4 | 1.8 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.4 | 1.3 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.4 | 0.9 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.4 | 2.1 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.4 | 5.4 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.4 | 1.2 | GO:0009624 | response to nematode(GO:0009624) |
| 0.4 | 8.1 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.4 | 2.4 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.4 | 14.8 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.4 | 5.9 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.4 | 1.5 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.4 | 2.2 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.4 | 3.4 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.4 | 6.2 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.4 | 2.5 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.4 | 1.8 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.3 | 2.0 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.3 | 3.0 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.3 | 0.3 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.3 | 3.6 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.3 | 5.2 | GO:0007567 | parturition(GO:0007567) |
| 0.3 | 4.8 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.3 | 3.5 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.3 | 1.3 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.3 | 25.0 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.3 | 0.6 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.3 | 7.4 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.3 | 0.9 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.3 | 3.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.3 | 1.1 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.3 | 4.3 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.3 | 2.3 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.3 | 1.7 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.3 | 4.5 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.3 | 1.4 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.3 | 2.5 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.3 | 3.5 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.3 | 1.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.3 | 0.8 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.3 | 2.1 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.3 | 3.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.3 | 1.0 | GO:1900228 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.2 | 5.9 | GO:0048026 | positive regulation of circadian rhythm(GO:0042753) positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 1.7 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.2 | 1.0 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.2 | 0.7 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.2 | 5.6 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.2 | 1.0 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.2 | 0.7 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.2 | 0.7 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.2 | 3.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.2 | 1.9 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.2 | 0.5 | GO:0003245 | growth involved in heart morphogenesis(GO:0003241) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.2 | 6.9 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.2 | 1.9 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.2 | 1.9 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.2 | 2.1 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.2 | 3.1 | GO:0006558 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.2 | 0.6 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.2 | 0.8 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.2 | 1.7 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.2 | 4.7 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.2 | 2.3 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.2 | 1.3 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.2 | 3.3 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.2 | 14.4 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.2 | 1.3 | GO:0052055 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.2 | 1.4 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.2 | 2.3 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.2 | 0.2 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.2 | 0.4 | GO:1900005 | positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.2 | 1.0 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.2 | 0.9 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.2 | 2.2 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.2 | 1.7 | GO:0003360 | brainstem development(GO:0003360) |
| 0.2 | 2.6 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 0.6 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.2 | 7.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.2 | 6.1 | GO:0006505 | GPI anchor metabolic process(GO:0006505) GPI anchor biosynthetic process(GO:0006506) |
| 0.2 | 2.9 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.2 | 2.0 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.2 | 4.1 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.2 | 4.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 2.1 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.1 | 0.6 | GO:0071638 | cellular triglyceride homeostasis(GO:0035356) negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.1 | 1.8 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 4.2 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 0.9 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.1 | 0.7 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 1.6 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 3.0 | GO:0046697 | decidualization(GO:0046697) |
| 0.1 | 0.8 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 0.6 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 6.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.7 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.1 | 1.7 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 2.3 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 0.5 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 2.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 3.0 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 4.7 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 6.3 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 0.4 | GO:0051821 | negative regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010768) transmission of virus(GO:0019089) dissemination or transmission of symbiont from host(GO:0044007) dissemination or transmission of organism from other organism involved in symbiotic interaction(GO:0051821) |
| 0.1 | 0.2 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.1 | 0.6 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 1.4 | GO:1901978 | positive regulation of cell cycle checkpoint(GO:1901978) |
| 0.1 | 0.5 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) leading strand elongation(GO:0006272) |
| 0.1 | 6.2 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 2.0 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 1.7 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.1 | 1.4 | GO:0051775 | response to redox state(GO:0051775) |
| 0.1 | 4.5 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 1.8 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.1 | 0.4 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 1.2 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.1 | 7.7 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 2.3 | GO:0046051 | UTP metabolic process(GO:0046051) |
| 0.1 | 2.4 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.1 | 5.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 1.5 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.1 | 5.4 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 0.8 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.1 | 8.4 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.4 | GO:0060355 | regulation of cell adhesion molecule production(GO:0060353) positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.1 | 2.4 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.1 | 0.6 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 3.8 | GO:1903318 | negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.1 | 2.0 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.9 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 1.2 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 10.5 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.1 | 1.8 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 0.3 | GO:0051710 | cytolysis by symbiont of host cells(GO:0001897) regulation of cytolysis in other organism(GO:0051710) |
| 0.1 | 2.5 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 | 0.4 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.1 | 4.3 | GO:0032760 | positive regulation of tumor necrosis factor production(GO:0032760) |
| 0.1 | 0.4 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.1 | 1.6 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.1 | 1.6 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 4.1 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.1 | 0.4 | GO:0098700 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 2.5 | GO:0035904 | aorta development(GO:0035904) |
| 0.1 | 0.3 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.1 | 0.7 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.8 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 1.5 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 2.7 | GO:0006026 | aminoglycan catabolic process(GO:0006026) |
| 0.1 | 1.6 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.1 | 1.6 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.1 | 0.6 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.1 | 3.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 3.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.8 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 0.3 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 4.0 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 0.5 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.2 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.1 | 1.8 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.1 | 1.7 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 2.1 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 4.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.9 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 1.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.8 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.2 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 5.4 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 5.2 | GO:0048839 | inner ear development(GO:0048839) |
| 0.0 | 0.3 | GO:0032754 | positive regulation of interleukin-5 production(GO:0032754) |
| 0.0 | 2.5 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.0 | 4.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 1.1 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.7 | GO:0090128 | regulation of synapse maturation(GO:0090128) positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 3.3 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 1.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 1.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.9 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 0.3 | GO:0061436 | regulation of water loss via skin(GO:0033561) establishment of skin barrier(GO:0061436) |
| 0.0 | 1.5 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 2.4 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 1.4 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.3 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 1.0 | GO:0061053 | somite development(GO:0061053) |
| 0.0 | 3.0 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 0.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 1.0 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.2 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 1.1 | GO:0007127 | meiosis I(GO:0007127) |
| 0.0 | 0.2 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 64.9 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 3.8 | 37.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 3.5 | 14.1 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 3.4 | 10.2 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 3.2 | 29.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 3.2 | 9.5 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 2.3 | 9.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 1.8 | 23.4 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 1.7 | 26.2 | GO:0030478 | actin cap(GO:0030478) |
| 1.7 | 8.6 | GO:0001652 | granular component(GO:0001652) |
| 1.5 | 12.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 1.3 | 5.3 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 1.3 | 15.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 1.2 | 3.7 | GO:0016938 | kinesin I complex(GO:0016938) |
| 1.1 | 3.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.1 | 8.5 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 1.0 | 6.7 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.8 | 4.7 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.7 | 3.5 | GO:0031673 | H zone(GO:0031673) |
| 0.7 | 5.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.7 | 137.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.6 | 2.4 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.6 | 14.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.6 | 2.8 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.6 | 5.5 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.5 | 8.6 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.5 | 2.7 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.5 | 1.4 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.5 | 6.5 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.5 | 7.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.5 | 5.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.4 | 3.5 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.4 | 2.5 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.4 | 6.9 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.4 | 5.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.3 | 6.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.3 | 4.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.3 | 2.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.3 | 2.5 | GO:0031466 | VCB complex(GO:0030891) Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.3 | 3.8 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.3 | 5.7 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 0.7 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.2 | 9.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 5.6 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.2 | 1.0 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.2 | 5.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.2 | 1.0 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.2 | 5.9 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.2 | 2.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 2.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.2 | 2.8 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.2 | 1.7 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.2 | 4.5 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 5.0 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 1.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 1.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 4.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 3.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 2.9 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 0.3 | GO:0000805 | X chromosome(GO:0000805) |
| 0.1 | 3.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 7.6 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 3.3 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 5.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 2.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.5 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 5.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 0.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 4.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.7 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.4 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 1.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 7.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 3.8 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 0.4 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.1 | 5.2 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 1.2 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.1 | 1.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 3.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 4.2 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 0.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 3.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 3.6 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 3.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 4.1 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 14.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.6 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 52.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 3.8 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 5.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.0 | GO:0071564 | SWI/SNF complex(GO:0016514) npBAF complex(GO:0071564) |
| 0.0 | 0.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 2.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 5.0 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 3.3 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 1.8 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 3.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 15.7 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 3.3 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 4.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.6 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 3.2 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.6 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 3.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.7 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 9.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.5 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.3 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 1.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 2.6 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.4 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 4.1 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 19.0 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.5 | 25.4 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 7.3 | 29.2 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 6.2 | 24.7 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 4.4 | 30.8 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 4.2 | 21.0 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 3.9 | 34.8 | GO:0045545 | syndecan binding(GO:0045545) |
| 2.8 | 64.9 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 2.6 | 10.5 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) |
| 2.6 | 7.7 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 2.3 | 7.0 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 1.9 | 5.7 | GO:0017129 | triglyceride binding(GO:0017129) |
| 1.9 | 5.7 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 1.6 | 28.2 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 1.5 | 6.0 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 1.4 | 4.1 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 1.4 | 2.7 | GO:0030305 | heparanase activity(GO:0030305) |
| 1.3 | 5.3 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 1.3 | 9.3 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 1.2 | 5.0 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 1.1 | 8.5 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 1.1 | 12.7 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 1.0 | 6.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 1.0 | 3.0 | GO:0047783 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.9 | 2.7 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.9 | 4.3 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.8 | 5.9 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.8 | 4.1 | GO:0030109 | HLA-B specific inhibitory MHC class I receptor activity(GO:0030109) |
| 0.8 | 22.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.8 | 9.7 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.8 | 6.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.8 | 3.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.7 | 7.4 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.7 | 8.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.7 | 4.1 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.6 | 7.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.6 | 5.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.6 | 1.8 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.6 | 1.8 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.6 | 2.3 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.6 | 8.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.6 | 3.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.5 | 6.0 | GO:0004568 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.5 | 0.5 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.5 | 5.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.5 | 2.6 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.5 | 1.6 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.5 | 3.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.5 | 14.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.5 | 7.4 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.5 | 6.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.5 | 2.3 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.4 | 1.3 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.4 | 3.4 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.4 | 52.6 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.4 | 5.5 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.4 | 2.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.4 | 2.2 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.4 | 3.0 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.4 | 1.5 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.4 | 1.8 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.4 | 2.5 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.3 | 2.4 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.3 | 1.4 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.3 | 4.7 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.3 | 0.3 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.3 | 3.9 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.3 | 1.0 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.3 | 34.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.3 | 5.8 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.3 | 1.6 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.3 | 2.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.3 | 1.5 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.3 | 1.7 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.3 | 1.7 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.3 | 8.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 2.5 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.3 | 3.5 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.3 | 7.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.3 | 2.9 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.3 | 1.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.3 | 1.3 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.3 | 1.5 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.3 | 2.0 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.3 | 6.8 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.3 | 1.8 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.2 | 1.7 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.2 | 2.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 66.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 1.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.2 | 5.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 1.8 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.2 | 4.0 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.2 | 0.4 | GO:0031779 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.2 | 8.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 2.0 | GO:0005549 | odorant binding(GO:0005549) |
| 0.2 | 9.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 2.7 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.2 | 1.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 5.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 0.7 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.2 | 7.1 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.2 | 2.0 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.2 | 4.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 2.8 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.2 | 1.6 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.2 | 9.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.7 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 0.9 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 1.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 1.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 4.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 3.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 11.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.7 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 3.1 | GO:0052744 | phosphatidylinositol-3-phosphatase activity(GO:0004438) phosphatidylinositol monophosphate phosphatase activity(GO:0052744) |
| 0.1 | 6.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 6.9 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 4.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 2.4 | GO:0034185 | triglyceride lipase activity(GO:0004806) apolipoprotein binding(GO:0034185) |
| 0.1 | 1.2 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 3.0 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 2.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 10.4 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.1 | 2.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 1.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 2.2 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 1.7 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 0.4 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 1.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 2.4 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.1 | 1.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.4 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.1 | 0.4 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 2.5 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 1.7 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.9 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 1.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 1.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 8.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 0.9 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 1.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 2.3 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 0.7 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.1 | 0.9 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.6 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 1.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 2.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.5 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.1 | 12.0 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.1 | 4.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.4 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.8 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 6.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 3.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.6 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 1.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.3 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 1.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 1.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 12.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.6 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 1.1 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 1.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.2 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 2.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 2.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.7 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 1.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.9 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.8 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.1 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.0 | 1.1 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.2 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.1 | GO:0015386 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.4 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 19.9 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.8 | 59.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.7 | 43.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.6 | 19.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.6 | 24.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.6 | 8.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.4 | 2.5 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.4 | 49.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.2 | 2.8 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.2 | 2.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.2 | 35.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 7.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.2 | 7.0 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 49.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 6.1 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.2 | 7.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 7.9 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.2 | 3.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 7.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 7.7 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 1.6 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 6.0 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 1.8 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 7.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 1.9 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 5.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 1.7 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 5.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 3.8 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 6.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 3.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 3.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 6.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 1.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 3.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 5.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 0.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 4.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 8.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 14.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.7 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.5 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.6 | NABA MATRISOME | Ensemble of genes encoding extracellular matrix and extracellular matrix-associated proteins |
| 0.0 | 0.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 51.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 2.1 | 27.0 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 1.7 | 19.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 1.0 | 22.0 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.9 | 23.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.6 | 30.4 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.6 | 5.7 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.5 | 28.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.5 | 66.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.5 | 17.6 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.4 | 52.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.4 | 3.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.4 | 5.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.4 | 7.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 14.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 5.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.3 | 6.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.3 | 7.7 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.3 | 24.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.3 | 6.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.3 | 5.6 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.3 | 1.9 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.3 | 7.9 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.3 | 2.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 5.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 7.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.2 | 2.4 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.2 | 5.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 39.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.2 | 6.3 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.2 | 3.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.2 | 2.0 | REACTOME OPSINS | Genes involved in Opsins |
| 0.2 | 9.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 3.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 3.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.2 | 1.8 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 5.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.2 | 5.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 9.9 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 7.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 2.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 3.0 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 2.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 2.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 4.1 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 1.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 1.0 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 2.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 2.3 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 8.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 5.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 0.7 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.1 | 0.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.5 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 6.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 2.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 2.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 0.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 1.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 4.4 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 1.8 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 4.8 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 0.5 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.7 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.6 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 2.0 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.5 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 1.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 3.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.2 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.0 | 0.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |