Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for TBP
Z-value: 2.54
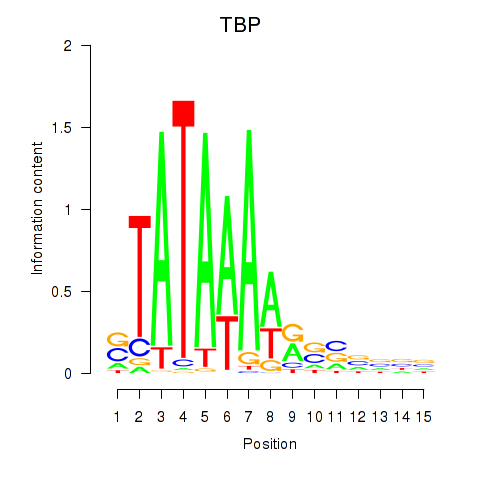
Transcription factors associated with TBP
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBP
|
ENSG00000112592.14 | TBP |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBP | hg38_v1_chr6_+_170554333_170554396 | -0.34 | 1.8e-07 | Click! |
Activity profile of TBP motif
Sorted Z-values of TBP motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TBP
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 20.3 | 61.0 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 16.3 | 65.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 16.1 | 16.1 | GO:1900154 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 14.9 | 44.8 | GO:0001796 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) |
| 12.4 | 49.7 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 10.3 | 10.3 | GO:1905237 | response to cyclosporin A(GO:1905237) |
| 9.7 | 38.9 | GO:0003431 | growth plate cartilage chondrocyte growth(GO:0003430) growth plate cartilage chondrocyte development(GO:0003431) |
| 9.5 | 9.5 | GO:0003129 | heart induction(GO:0003129) |
| 7.5 | 22.4 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 7.2 | 50.1 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 6.9 | 13.8 | GO:0045168 | cell-cell signaling involved in cell fate commitment(GO:0045168) |
| 6.8 | 27.3 | GO:0070426 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 6.4 | 19.1 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 6.3 | 25.0 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 6.0 | 30.1 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 6.0 | 143.9 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 5.9 | 105.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 5.8 | 17.4 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 5.8 | 23.1 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 5.8 | 17.3 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 5.6 | 72.8 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 5.6 | 28.0 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 5.3 | 26.6 | GO:0010046 | response to mycotoxin(GO:0010046) iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 5.3 | 15.8 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 5.2 | 26.0 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 4.9 | 19.6 | GO:1902617 | response to fluoride(GO:1902617) |
| 4.8 | 28.6 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 4.7 | 14.0 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 4.6 | 9.2 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 4.6 | 22.9 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 4.5 | 35.8 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 4.4 | 13.3 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 4.0 | 15.9 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 3.7 | 14.9 | GO:1900228 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 3.7 | 11.1 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 3.6 | 10.9 | GO:0001207 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 3.5 | 17.5 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 3.5 | 10.4 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 3.5 | 17.3 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 3.3 | 30.0 | GO:1902255 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) fat pad development(GO:0060613) positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) positive regulation of protein monoubiquitination(GO:1902527) |
| 3.3 | 9.9 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 3.1 | 6.3 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 3.1 | 9.4 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 3.1 | 9.3 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 3.0 | 12.0 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 2.8 | 11.3 | GO:0070295 | renal water absorption(GO:0070295) |
| 2.8 | 19.6 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 2.8 | 5.6 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 2.8 | 16.6 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 2.7 | 16.5 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 2.7 | 13.7 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 2.7 | 8.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 2.7 | 29.7 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 2.7 | 8.1 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 2.5 | 12.6 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 2.5 | 7.5 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 2.5 | 14.8 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 2.5 | 14.7 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 2.4 | 9.5 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 2.3 | 57.3 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 2.3 | 43.0 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 2.3 | 15.8 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 2.1 | 8.4 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 2.0 | 6.1 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 2.0 | 14.0 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 2.0 | 58.5 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 2.0 | 5.9 | GO:0006533 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 1.9 | 5.8 | GO:0015847 | putrescine transport(GO:0015847) |
| 1.9 | 5.7 | GO:0001983 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 1.9 | 5.6 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 1.8 | 19.7 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 1.8 | 10.7 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 1.7 | 12.2 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 1.7 | 8.7 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 1.7 | 10.4 | GO:0060356 | leucine import(GO:0060356) |
| 1.7 | 66.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 1.7 | 10.3 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 1.7 | 20.3 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 1.7 | 8.4 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 1.6 | 6.6 | GO:0035962 | response to interleukin-13(GO:0035962) |
| 1.6 | 14.7 | GO:0097267 | omega-hydroxylase P450 pathway(GO:0097267) |
| 1.6 | 29.4 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 1.6 | 4.7 | GO:0035993 | subthalamic nucleus development(GO:0021763) deltoid tuberosity development(GO:0035993) prolactin secreting cell differentiation(GO:0060127) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 1.6 | 40.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 1.6 | 12.5 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 1.6 | 10.9 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 1.5 | 6.2 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 1.5 | 9.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 1.5 | 6.0 | GO:0035106 | operant conditioning(GO:0035106) |
| 1.5 | 4.4 | GO:0060032 | notochord regression(GO:0060032) |
| 1.4 | 11.5 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 1.4 | 4.3 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 1.4 | 28.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 1.4 | 5.6 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 1.4 | 4.1 | GO:0015669 | gas transport(GO:0015669) |
| 1.4 | 4.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 1.4 | 13.6 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 1.4 | 9.5 | GO:0032097 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 1.4 | 6.8 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 1.3 | 9.2 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 1.3 | 1.3 | GO:1904437 | positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 1.3 | 2.6 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 1.3 | 3.8 | GO:2000848 | positive regulation of corticosteroid hormone secretion(GO:2000848) |
| 1.3 | 8.9 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 1.3 | 2.5 | GO:0014806 | smooth muscle hyperplasia(GO:0014806) |
| 1.3 | 6.3 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 1.2 | 4.9 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 1.2 | 35.4 | GO:0001502 | cartilage condensation(GO:0001502) |
| 1.2 | 5.8 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 1.2 | 12.7 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 1.1 | 50.2 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 1.1 | 29.3 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 1.1 | 8.8 | GO:0038030 | non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 1.1 | 4.4 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 1.1 | 5.4 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 1.0 | 3.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 1.0 | 32.2 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 1.0 | 4.1 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 1.0 | 12.1 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 1.0 | 10.8 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 1.0 | 3.9 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 1.0 | 13.8 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 1.0 | 17.5 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 1.0 | 6.8 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 1.0 | 3.8 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.9 | 12.2 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.9 | 2.7 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.9 | 15.4 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.9 | 3.4 | GO:0002357 | defense response to tumor cell(GO:0002357) |
| 0.9 | 33.3 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.8 | 2.5 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.8 | 9.0 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.8 | 19.5 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.8 | 3.2 | GO:0070092 | glucagon secretion(GO:0070091) regulation of glucagon secretion(GO:0070092) |
| 0.8 | 7.9 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.8 | 3.9 | GO:0032667 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) positive regulation of interleukin-23 production(GO:0032747) |
| 0.8 | 23.8 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.8 | 10.8 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.7 | 12.7 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.7 | 41.9 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.7 | 7.9 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.7 | 1.4 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.7 | 10.3 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.6 | 2.6 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.6 | 3.8 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.6 | 12.0 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.6 | 4.4 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.6 | 49.1 | GO:0070268 | cornification(GO:0070268) |
| 0.6 | 4.2 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.6 | 16.8 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.6 | 4.1 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.6 | 4.1 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.6 | 5.7 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.5 | 2.7 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.5 | 5.8 | GO:0030728 | ovulation(GO:0030728) |
| 0.5 | 17.7 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.5 | 2.1 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.5 | 1.0 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.5 | 29.5 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.5 | 2.4 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.5 | 7.8 | GO:0031424 | keratinization(GO:0031424) |
| 0.5 | 1.8 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.5 | 1.4 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.5 | 11.0 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.4 | 6.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.4 | 6.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.4 | 12.0 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.4 | 9.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.4 | 2.0 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.4 | 1.5 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.4 | 1.1 | GO:0021780 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.3 | 1.4 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.3 | 1.7 | GO:2000858 | mineralocorticoid secretion(GO:0035931) aldosterone secretion(GO:0035932) regulation of mineralocorticoid secretion(GO:2000855) regulation of aldosterone secretion(GO:2000858) |
| 0.3 | 1.9 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.3 | 4.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.3 | 6.8 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.3 | 1.8 | GO:0001692 | histamine metabolic process(GO:0001692) histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.3 | 5.0 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.3 | 2.3 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.3 | 0.8 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.2 | 9.2 | GO:0034694 | response to prostaglandin(GO:0034694) |
| 0.2 | 11.2 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.2 | 23.8 | GO:0007586 | digestion(GO:0007586) |
| 0.2 | 1.1 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.2 | 5.3 | GO:1903206 | negative regulation of response to reactive oxygen species(GO:1901032) negative regulation of hydrogen peroxide-induced cell death(GO:1903206) |
| 0.2 | 4.8 | GO:0007530 | sex determination(GO:0007530) |
| 0.2 | 5.8 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.2 | 2.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 15.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.2 | 12.1 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.2 | 6.6 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.2 | 4.3 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.2 | 10.2 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.2 | 8.8 | GO:2000273 | positive regulation of receptor activity(GO:2000273) |
| 0.2 | 11.5 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.2 | 5.1 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.2 | 15.2 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.2 | 2.7 | GO:0072678 | T cell migration(GO:0072678) |
| 0.2 | 20.5 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.2 | 1.7 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.2 | 1.7 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.2 | 0.6 | GO:0009305 | protein biotinylation(GO:0009305) histone biotinylation(GO:0071110) |
| 0.1 | 1.5 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.1 | 0.7 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 1.8 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.1 | 1.1 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.1 | 6.1 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 12.2 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 4.9 | GO:1904031 | positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 | 4.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 10.1 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 0.5 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.1 | 2.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 17.0 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.1 | 3.8 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.1 | 2.4 | GO:0060384 | response to histamine(GO:0034776) innervation(GO:0060384) cellular response to histamine(GO:0071420) |
| 0.1 | 0.3 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.1 | 5.4 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.1 | 2.7 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 5.6 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 0.1 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.1 | 4.1 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.1 | 4.7 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.1 | 0.9 | GO:0014911 | positive regulation of smooth muscle cell migration(GO:0014911) |
| 0.0 | 1.4 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 3.3 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.3 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.6 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.4 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 5.6 | GO:0010952 | positive regulation of peptidase activity(GO:0010952) |
| 0.0 | 2.2 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 1.2 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 4.6 | GO:0043542 | endothelial cell migration(GO:0043542) |
| 0.0 | 0.8 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 2.8 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 1.7 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 1.7 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 1.3 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 | 1.7 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 0.1 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 1.7 | GO:0034332 | adherens junction organization(GO:0034332) |
| 0.0 | 2.3 | GO:0009749 | response to glucose(GO:0009749) |
| 0.0 | 1.6 | GO:0042445 | hormone metabolic process(GO:0042445) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.2 | 61.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 14.3 | 43.0 | GO:0097229 | sperm end piece(GO:0097229) |
| 8.1 | 105.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 7.8 | 23.5 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 5.6 | 72.8 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 5.1 | 15.4 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 4.0 | 44.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 4.0 | 27.9 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 2.6 | 49.8 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 2.6 | 12.8 | GO:0036128 | CatSper complex(GO:0036128) |
| 2.3 | 9.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 2.2 | 44.6 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 2.1 | 59.6 | GO:0097386 | glial cell projection(GO:0097386) |
| 2.1 | 16.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 2.0 | 9.9 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 1.8 | 44.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 1.8 | 79.1 | GO:0045095 | keratin filament(GO:0045095) |
| 1.7 | 12.1 | GO:1990745 | EARP complex(GO:1990745) |
| 1.5 | 49.7 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 1.5 | 19.6 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 1.5 | 68.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 1.3 | 15.3 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 1.2 | 13.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 1.2 | 8.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 1.2 | 9.2 | GO:0045179 | apical cortex(GO:0045179) |
| 1.2 | 105.0 | GO:0035580 | specific granule lumen(GO:0035580) |
| 1.1 | 20.5 | GO:0036038 | MKS complex(GO:0036038) |
| 1.1 | 6.8 | GO:0072687 | meiotic spindle(GO:0072687) |
| 1.1 | 4.4 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 1.0 | 7.3 | GO:0001939 | female pronucleus(GO:0001939) |
| 1.0 | 50.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 1.0 | 17.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 1.0 | 28.9 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.9 | 4.5 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.9 | 6.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.9 | 4.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.9 | 37.5 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.7 | 145.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.7 | 2.7 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.6 | 4.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.6 | 4.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.5 | 5.8 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.5 | 37.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.5 | 4.6 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.5 | 1.4 | GO:0034677 | integrin alpha7-beta1 complex(GO:0034677) |
| 0.4 | 2.6 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.4 | 4.7 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.4 | 11.8 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.4 | 5.8 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.4 | 21.1 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.4 | 1.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.3 | 12.2 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.3 | 4.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.3 | 27.6 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.3 | 487.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.3 | 2.4 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.3 | 10.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.3 | 19.9 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.3 | 49.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.3 | 10.4 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.2 | 8.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 0.8 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.2 | 5.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.2 | 3.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.2 | 11.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 12.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.2 | 130.9 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 3.0 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 11.1 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 4.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 4.4 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.6 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 3.2 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 13.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 5.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 6.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 11.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 2.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.5 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 34.6 | GO:0009986 | cell surface(GO:0009986) |
| 0.1 | 6.1 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.8 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 1.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 3.7 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 1.3 | GO:0030496 | midbody(GO:0030496) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.3 | 65.1 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 12.2 | 36.6 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 11.1 | 166.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 9.9 | 29.7 | GO:0035375 | zymogen binding(GO:0035375) |
| 7.7 | 23.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 6.0 | 30.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 5.3 | 15.8 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 4.9 | 19.8 | GO:0035473 | lipase binding(GO:0035473) |
| 4.7 | 14.0 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 4.0 | 60.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 3.9 | 15.5 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 3.7 | 44.6 | GO:0008430 | selenium binding(GO:0008430) |
| 3.6 | 10.8 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 3.6 | 14.2 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 3.5 | 55.3 | GO:0019841 | retinol binding(GO:0019841) |
| 3.4 | 10.3 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 3.4 | 10.3 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 2.8 | 11.3 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 2.7 | 18.8 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 2.7 | 10.7 | GO:0045569 | TRAIL binding(GO:0045569) |
| 2.5 | 50.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 2.5 | 27.3 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 2.5 | 14.7 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 2.4 | 7.3 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 2.4 | 7.2 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 2.3 | 11.5 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 2.3 | 11.3 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 2.0 | 14.0 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 2.0 | 5.9 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 1.9 | 5.8 | GO:0015203 | polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) |
| 1.8 | 11.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 1.8 | 5.4 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 1.8 | 19.6 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 1.8 | 12.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 1.8 | 8.8 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 1.7 | 8.7 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 1.7 | 5.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 1.6 | 6.6 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 1.6 | 6.3 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 1.6 | 12.6 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 1.5 | 7.5 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 1.5 | 43.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 1.4 | 5.7 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 1.4 | 35.5 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 1.4 | 10.9 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 1.2 | 3.7 | GO:0008903 | hydroxypyruvate isomerase activity(GO:0008903) |
| 1.2 | 17.3 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 1.2 | 155.6 | GO:0005179 | hormone activity(GO:0005179) |
| 1.2 | 6.1 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 1.2 | 39.7 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 1.1 | 5.7 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 1.1 | 4.4 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 1.1 | 5.4 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 1.0 | 85.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 1.0 | 12.5 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 1.0 | 10.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 1.0 | 14.5 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 1.0 | 7.1 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 1.0 | 17.2 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 1.0 | 4.9 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 1.0 | 7.8 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 1.0 | 8.6 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) phosphate ion binding(GO:0042301) |
| 0.9 | 22.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.9 | 6.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.9 | 25.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.8 | 4.2 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.8 | 9.0 | GO:0008061 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.8 | 10.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.7 | 78.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.7 | 5.8 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.7 | 1.4 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.7 | 2.8 | GO:0004773 | steryl-sulfatase activity(GO:0004773) |
| 0.7 | 12.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.7 | 6.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.7 | 8.2 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.7 | 5.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.7 | 32.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.6 | 17.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.6 | 10.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.6 | 5.0 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.6 | 3.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.6 | 16.8 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.6 | 19.7 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.6 | 2.4 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.6 | 5.6 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.5 | 5.8 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.5 | 6.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.5 | 140.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.5 | 2.5 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.5 | 2.0 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.5 | 40.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.5 | 10.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.5 | 3.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.5 | 1.4 | GO:0005135 | interleukin-3 receptor binding(GO:0005135) |
| 0.5 | 1.8 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.4 | 56.5 | GO:0008201 | heparin binding(GO:0008201) |
| 0.4 | 4.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.4 | 12.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.4 | 13.1 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.4 | 1.6 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.4 | 10.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.4 | 8.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.4 | 2.7 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.4 | 8.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.4 | 3.7 | GO:0005549 | odorant binding(GO:0005549) |
| 0.3 | 4.1 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.3 | 10.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.3 | 17.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.3 | 4.9 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.3 | 4.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.3 | 4.7 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.3 | 1.6 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.3 | 2.4 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.3 | 7.4 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.3 | 10.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.3 | 1.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.3 | 10.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.3 | 2.7 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.3 | 2.8 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.3 | 2.5 | GO:0015386 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.3 | 4.0 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.3 | 6.8 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 3.2 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.2 | 2.4 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.2 | 43.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.2 | 4.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.2 | 22.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.2 | 1.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.2 | 24.6 | GO:0002020 | protease binding(GO:0002020) |
| 0.2 | 1.0 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.2 | 4.2 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.2 | 1.7 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.2 | 20.0 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.2 | 8.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.2 | 1.5 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 21.0 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.2 | 5.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 4.3 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.2 | 17.5 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.2 | 4.3 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.2 | 4.7 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.2 | 13.8 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.2 | 7.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.2 | 0.6 | GO:0018271 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.1 | 2.0 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 5.4 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.4 | GO:0030547 | receptor inhibitor activity(GO:0030547) |
| 0.1 | 110.4 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.1 | 0.3 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.1 | 0.8 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 6.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.8 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.1 | 1.3 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 1.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 5.6 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 10.7 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 4.9 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 2.2 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 4.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 4.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 0.5 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.1 | 45.3 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.1 | 0.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 4.1 | GO:0016782 | transferase activity, transferring sulfur-containing groups(GO:0016782) |
| 0.0 | 1.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 8.0 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 1.1 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 3.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 13.9 | GO:0000987 | core promoter proximal region sequence-specific DNA binding(GO:0000987) |
| 0.0 | 2.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 7.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.8 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 5.8 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 12.4 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 3.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.2 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.7 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 136.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 1.6 | 44.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 1.4 | 49.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 1.4 | 5.6 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 1.0 | 56.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.9 | 12.5 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.8 | 42.8 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.8 | 19.6 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.7 | 28.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.7 | 28.3 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.6 | 18.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.6 | 44.9 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.5 | 18.6 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.5 | 18.9 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.5 | 11.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.5 | 171.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.4 | 23.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.4 | 16.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.4 | 115.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.4 | 10.7 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.4 | 15.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.4 | 22.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.3 | 32.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.3 | 2.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.3 | 9.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.3 | 50.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 9.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 5.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 13.3 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.2 | 10.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.2 | 12.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.2 | 10.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.2 | 7.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.2 | 10.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.2 | 35.3 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.2 | 11.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.2 | 3.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.2 | 19.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.2 | 4.7 | ST ADRENERGIC | Adrenergic Pathway |
| 0.1 | 5.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 9.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 4.2 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 5.8 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 6.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 2.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 1.7 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 4.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 5.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 2.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 2.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.7 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 53.7 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 2.7 | 72.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 2.4 | 67.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 2.2 | 41.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 2.1 | 46.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 1.6 | 15.8 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 1.5 | 33.7 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 1.4 | 55.4 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 1.3 | 21.5 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 1.2 | 21.0 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 1.2 | 13.2 | REACTOME DEFENSINS | Genes involved in Defensins |
| 1.2 | 29.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 1.1 | 46.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 1.0 | 12.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.9 | 32.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.9 | 12.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.8 | 14.7 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.8 | 159.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.8 | 12.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.8 | 17.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.7 | 16.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.7 | 106.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.6 | 67.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.6 | 23.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.6 | 7.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.5 | 11.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.4 | 2.8 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.3 | 33.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.3 | 3.7 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.3 | 2.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 4.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.3 | 13.8 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.3 | 2.5 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 2.8 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 5.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 10.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 10.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 4.9 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.2 | 7.6 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.2 | 7.7 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.2 | 11.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.2 | 15.9 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.2 | 4.1 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 9.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 3.0 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 6.0 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 6.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 5.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 7.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.3 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 0.8 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.1 | 8.0 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.1 | 5.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 8.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 4.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 4.9 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 1.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 2.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 2.4 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 1.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.5 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 3.7 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 1.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |