|
chr14_-_106185387
Show fit
|
20.99 |
ENST00000390605.2
|
IGHV1-18
|
immunoglobulin heavy variable 1-18
|
|
chrX_+_120604084
Show fit
|
19.75 |
ENST00000371317.10
|
MCTS1
|
MCTS1 re-initiation and release factor
|
|
chr16_+_84768246
Show fit
|
18.96 |
ENST00000569925.1
ENST00000567526.1
|
USP10
|
ubiquitin specific peptidase 10
|
|
chr6_-_36547400
Show fit
|
18.74 |
ENST00000229812.8
|
STK38
|
serine/threonine kinase 38
|
|
chr9_-_113410666
Show fit
|
18.65 |
ENST00000374171.5
|
POLE3
|
DNA polymerase epsilon 3, accessory subunit
|
|
chr10_-_119536533
Show fit
|
18.45 |
ENST00000392865.5
|
RGS10
|
regulator of G protein signaling 10
|
|
chrX_+_120604199
Show fit
|
16.03 |
ENST00000371315.3
|
MCTS1
|
MCTS1 re-initiation and release factor
|
|
chr15_+_41286011
Show fit
|
15.76 |
ENST00000661438.1
|
ENSG00000285920.2
|
novel protein
|
|
chr2_-_85409805
Show fit
|
15.18 |
ENST00000449030.5
|
CAPG
|
capping actin protein, gelsolin like
|
|
chr10_-_93482326
Show fit
|
15.03 |
ENST00000359263.9
|
MYOF
|
myoferlin
|
|
chr10_-_93482194
Show fit
|
14.59 |
ENST00000358334.9
ENST00000371488.3
|
MYOF
|
myoferlin
|
|
chr7_-_133082032
Show fit
|
14.50 |
ENST00000448878.6
ENST00000262570.10
|
CHCHD3
|
coiled-coil-helix-coiled-coil-helix domain containing 3
|
|
chr6_-_2841853
Show fit
|
14.10 |
ENST00000380739.6
|
SERPINB1
|
serpin family B member 1
|
|
chr7_-_148883474
Show fit
|
13.86 |
ENST00000476773.5
|
EZH2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit
|
|
chr12_-_48925483
Show fit
|
13.37 |
ENST00000550765.6
ENST00000552878.5
ENST00000453172.2
|
FKBP11
|
FKBP prolyl isomerase 11
|
|
chr1_+_32292067
Show fit
|
13.27 |
ENST00000373548.8
ENST00000428704.1
|
HDAC1
|
histone deacetylase 1
|
|
chr7_+_2354810
Show fit
|
12.84 |
ENST00000360876.9
ENST00000413917.5
ENST00000397011.2
|
EIF3B
|
eukaryotic translation initiation factor 3 subunit B
|
|
chr12_+_52069967
Show fit
|
12.60 |
ENST00000336854.9
ENST00000550604.1
ENST00000553049.5
ENST00000548915.1
|
ATG101
|
autophagy related 101
|
|
chr13_+_50909905
Show fit
|
12.20 |
ENST00000644034.1
ENST00000645955.1
|
RNASEH2B
|
ribonuclease H2 subunit B
|
|
chr14_-_106507476
Show fit
|
12.15 |
ENST00000390621.3
|
IGHV1-45
|
immunoglobulin heavy variable 1-45
|
|
chr1_-_160647287
Show fit
|
12.13 |
ENST00000235739.6
|
SLAMF1
|
signaling lymphocytic activation molecule family member 1
|
|
chr8_-_63038788
Show fit
|
11.87 |
ENST00000518113.2
ENST00000260118.7
ENST00000677482.1
|
GGH
|
gamma-glutamyl hydrolase
|
|
chr10_-_119178791
Show fit
|
11.72 |
ENST00000298510.4
|
PRDX3
|
peroxiredoxin 3
|
|
chr15_-_55289756
Show fit
|
11.45 |
ENST00000336787.6
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr11_+_72227881
Show fit
|
10.97 |
ENST00000538751.5
ENST00000541756.5
|
INPPL1
|
inositol polyphosphate phosphatase like 1
|
|
chr1_+_158930778
Show fit
|
10.73 |
ENST00000458222.5
|
PYHIN1
|
pyrin and HIN domain family member 1
|
|
chr1_-_63523175
Show fit
|
10.71 |
ENST00000371092.7
ENST00000271002.15
|
ITGB3BP
|
integrin subunit beta 3 binding protein
|
|
chr14_+_20781139
Show fit
|
10.58 |
ENST00000304677.3
|
RNASE6
|
ribonuclease A family member k6
|
|
chr2_+_90114838
Show fit
|
10.55 |
ENST00000417279.3
|
IGKV3D-15
|
immunoglobulin kappa variable 3D-15
|
|
chr1_+_171512032
Show fit
|
10.09 |
ENST00000426496.6
|
PRRC2C
|
proline rich coiled-coil 2C
|
|
chr2_+_200889327
Show fit
|
9.76 |
ENST00000426253.5
ENST00000454952.1
ENST00000651500.1
ENST00000409020.6
ENST00000359683.8
|
NIF3L1
|
NGG1 interacting factor 3 like 1
|
|
chr5_+_96876480
Show fit
|
9.60 |
ENST00000437043.8
ENST00000379904.8
|
ERAP2
|
endoplasmic reticulum aminopeptidase 2
|
|
chr5_+_33440696
Show fit
|
9.46 |
ENST00000502553.5
ENST00000514259.5
|
TARS1
|
threonyl-tRNA synthetase 1
|
|
chr6_+_22569554
Show fit
|
9.38 |
ENST00000510882.4
|
HDGFL1
|
HDGF like 1
|
|
chr1_-_150697128
Show fit
|
9.31 |
ENST00000427665.1
ENST00000271732.8
|
GOLPH3L
|
golgi phosphoprotein 3 like
|
|
chr1_-_193105373
Show fit
|
8.76 |
ENST00000367439.8
|
GLRX2
|
glutaredoxin 2
|
|
chr14_+_23988884
Show fit
|
8.71 |
ENST00000558753.5
ENST00000537912.5
|
DHRS4L2
|
dehydrogenase/reductase 4 like 2
|
|
chr2_+_200889411
Show fit
|
8.69 |
ENST00000409357.5
ENST00000409129.2
|
NIF3L1
|
NGG1 interacting factor 3 like 1
|
|
chr2_-_89027700
Show fit
|
8.59 |
ENST00000483158.1
|
IGKV3-11
|
immunoglobulin kappa variable 3-11
|
|
chr10_+_75210151
Show fit
|
8.50 |
ENST00000298468.9
ENST00000543351.5
|
VDAC2
|
voltage dependent anion channel 2
|
|
chr1_-_160647037
Show fit
|
8.38 |
ENST00000302035.11
|
SLAMF1
|
signaling lymphocytic activation molecule family member 1
|
|
chr1_+_207454230
Show fit
|
8.28 |
ENST00000367058.7
ENST00000367059.3
ENST00000367057.8
|
CR2
|
complement C3d receptor 2
|
|
chr2_-_27323006
Show fit
|
7.87 |
ENST00000402310.5
ENST00000405983.5
ENST00000403262.6
|
MPV17
|
mitochondrial inner membrane protein MPV17
|
|
chr22_+_44677044
Show fit
|
7.79 |
ENST00000006251.11
|
PRR5
|
proline rich 5
|
|
chr7_-_141014939
Show fit
|
7.54 |
ENST00000324787.10
ENST00000467334.1
|
MRPS33
|
mitochondrial ribosomal protein S33
|
|
chr2_+_89851723
Show fit
|
7.35 |
ENST00000429992.2
|
IGKV2D-40
|
immunoglobulin kappa variable 2D-40
|
|
chr19_+_48993525
Show fit
|
7.05 |
ENST00000601968.5
ENST00000596837.5
|
RUVBL2
|
RuvB like AAA ATPase 2
|
|
chr6_+_52420107
Show fit
|
6.99 |
ENST00000636489.1
ENST00000637089.1
ENST00000637353.1
ENST00000637263.1
|
EFHC1
|
EF-hand domain containing 1
|
|
chr17_-_64505357
Show fit
|
6.97 |
ENST00000583212.2
ENST00000578190.5
ENST00000579091.5
ENST00000583239.6
|
DDX5
|
DEAD-box helicase 5
|
|
chr5_+_36166556
Show fit
|
6.81 |
ENST00000677886.1
|
SKP2
|
S-phase kinase associated protein 2
|
|
chr22_+_44677077
Show fit
|
6.61 |
ENST00000403581.5
|
PRR5
|
proline rich 5
|
|
chr5_+_35856883
Show fit
|
6.46 |
ENST00000506850.5
ENST00000303115.8
ENST00000511982.1
|
IL7R
|
interleukin 7 receptor
|
|
chr10_+_17644126
Show fit
|
6.40 |
ENST00000377524.8
|
STAM
|
signal transducing adaptor molecule
|
|
chr2_+_3575250
Show fit
|
6.14 |
ENST00000645674.2
|
RPS7
|
ribosomal protein S7
|
|
chr19_-_38847423
Show fit
|
6.08 |
ENST00000647557.1
|
HNRNPL
|
heterogeneous nuclear ribonucleoprotein L
|
|
chr20_+_3209469
Show fit
|
5.98 |
ENST00000380113.8
ENST00000455664.6
ENST00000399838.3
|
ITPA
|
inosine triphosphatase
|
|
chr2_-_88979016
Show fit
|
5.88 |
ENST00000390247.2
|
IGKV3-7
|
immunoglobulin kappa variable 3-7 (non-functional)
|
|
chr6_+_10694916
Show fit
|
5.72 |
ENST00000379568.4
|
PAK1IP1
|
PAK1 interacting protein 1
|
|
chr9_-_128127711
Show fit
|
5.70 |
ENST00000449878.1
ENST00000338961.11
ENST00000678174.1
|
PTGES2
|
prostaglandin E synthase 2
|
|
chr2_+_90234809
Show fit
|
5.64 |
ENST00000443397.5
|
IGKV3D-7
|
immunoglobulin kappa variable 3D-7
|
|
chr1_-_167935987
Show fit
|
5.56 |
ENST00000367846.8
|
MPC2
|
mitochondrial pyruvate carrier 2
|
|
chr1_-_150974867
Show fit
|
5.52 |
ENST00000271688.10
|
CERS2
|
ceramide synthase 2
|
|
chr2_+_143129379
Show fit
|
5.48 |
ENST00000295095.11
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chr6_+_52420332
Show fit
|
5.46 |
ENST00000636107.1
ENST00000371068.11
ENST00000636702.1
ENST00000635996.1
ENST00000636379.1
|
EFHC1
|
EF-hand domain containing 1
|
|
chr6_-_42746054
Show fit
|
5.28 |
ENST00000372876.2
|
TBCC
|
tubulin folding cofactor C
|
|
chr14_-_106154113
Show fit
|
5.18 |
ENST00000390603.2
|
IGHV3-15
|
immunoglobulin heavy variable 3-15
|
|
chr19_-_7702124
Show fit
|
4.78 |
ENST00000597921.6
|
FCER2
|
Fc fragment of IgE receptor II
|
|
chrX_-_120575783
Show fit
|
4.73 |
ENST00000680673.1
|
CUL4B
|
cullin 4B
|
|
chr17_+_7219857
Show fit
|
4.66 |
ENST00000583312.5
ENST00000350303.9
ENST00000584103.5
ENST00000356839.10
ENST00000579886.2
|
ACADVL
|
acyl-CoA dehydrogenase very long chain
|
|
chr7_-_76626127
Show fit
|
4.59 |
ENST00000454397.1
|
POMZP3
|
POM121 and ZP3 fusion
|
|
chr20_-_5113032
Show fit
|
4.59 |
ENST00000379299.6
ENST00000379286.6
ENST00000379279.6
ENST00000379283.6
|
TMEM230
|
transmembrane protein 230
|
|
chr17_+_36948925
Show fit
|
4.53 |
ENST00000616434.2
ENST00000680340.1
ENST00000619387.5
ENST00000679997.1
|
AATF
|
apoptosis antagonizing transcription factor
|
|
chr12_-_9607903
Show fit
|
4.35 |
ENST00000229402.4
|
KLRB1
|
killer cell lectin like receptor B1
|
|
chr19_-_7702139
Show fit
|
4.28 |
ENST00000346664.9
|
FCER2
|
Fc fragment of IgE receptor II
|
|
chr11_+_57657736
Show fit
|
4.27 |
ENST00000529773.2
ENST00000533905.1
ENST00000525602.1
ENST00000533682.2
ENST00000302731.4
|
CLP1
|
cleavage factor polyribonucleotide kinase subunit 1
|
|
chr17_-_78874038
Show fit
|
4.12 |
ENST00000586057.5
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr20_-_5113067
Show fit
|
4.09 |
ENST00000342308.10
ENST00000612323.4
ENST00000202834.11
|
TMEM230
|
transmembrane protein 230
|
|
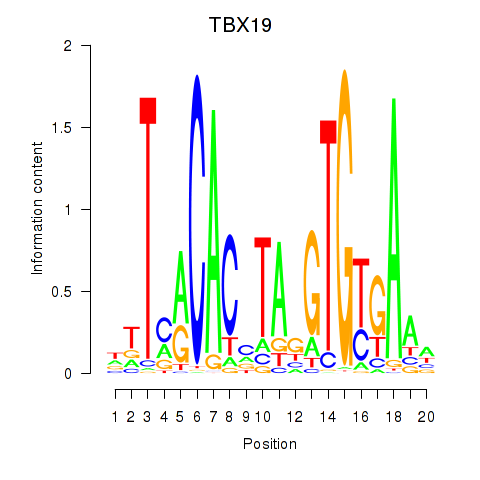
chr1_+_168280872
Show fit
|
4.06 |
ENST00000367821.8
|
TBX19
|
T-box transcription factor 19
|
|
chr10_+_5094405
Show fit
|
3.99 |
ENST00000380554.5
|
AKR1C3
|
aldo-keto reductase family 1 member C3
|
|
chr20_-_46363174
Show fit
|
3.99 |
ENST00000372227.5
|
SLC35C2
|
solute carrier family 35 member C2
|
|
chr18_+_62539511
Show fit
|
3.97 |
ENST00000586834.1
|
ZCCHC2
|
zinc finger CCHC-type containing 2
|
|
chr1_-_167553745
Show fit
|
3.90 |
ENST00000370509.5
|
CREG1
|
cellular repressor of E1A stimulated genes 1
|
|
chr15_-_60479080
Show fit
|
3.88 |
ENST00000560072.5
ENST00000560406.5
ENST00000560520.1
ENST00000261520.9
|
ICE2
|
interactor of little elongation complex ELL subunit 2
|
|
chr5_+_169583636
Show fit
|
3.77 |
ENST00000506574.5
ENST00000515224.5
ENST00000629457.2
ENST00000508247.5
ENST00000265295.9
ENST00000513941.5
|
SPDL1
|
spindle apparatus coiled-coil protein 1
|
|
chr1_+_116754422
Show fit
|
3.71 |
ENST00000369478.4
ENST00000369477.1
|
CD2
|
CD2 molecule
|
|
chr22_+_35257452
Show fit
|
3.69 |
ENST00000420166.5
ENST00000216106.6
ENST00000455359.5
|
HMGXB4
|
HMG-box containing 4
|
|
chr3_-_194672175
Show fit
|
3.60 |
ENST00000265245.10
|
LSG1
|
large 60S subunit nuclear export GTPase 1
|
|
chr14_-_106875069
Show fit
|
3.60 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional)
|
|
chr12_+_8123609
Show fit
|
3.52 |
ENST00000229332.12
|
CLEC4A
|
C-type lectin domain family 4 member A
|
|
chr2_-_9423340
Show fit
|
3.44 |
ENST00000484735.5
ENST00000456913.6
|
ITGB1BP1
|
integrin subunit beta 1 binding protein 1
|
|
chr1_+_113929304
Show fit
|
3.37 |
ENST00000426820.7
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr19_-_7943648
Show fit
|
3.33 |
ENST00000597926.1
ENST00000270538.8
|
TIMM44
|
translocase of inner mitochondrial membrane 44
|
|
chr1_+_113929350
Show fit
|
3.23 |
ENST00000369559.8
ENST00000626993.2
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr14_-_106511856
Show fit
|
3.12 |
ENST00000390622.2
|
IGHV1-46
|
immunoglobulin heavy variable 1-46
|
|
chr6_+_30617825
Show fit
|
3.12 |
ENST00000259873.5
|
MRPS18B
|
mitochondrial ribosomal protein S18B
|
|
chr17_-_19745369
Show fit
|
3.10 |
ENST00000573368.5
ENST00000457500.6
|
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1
|
|
chr2_-_112836702
Show fit
|
3.07 |
ENST00000416750.1
ENST00000263341.7
ENST00000418817.5
|
IL1B
|
interleukin 1 beta
|
|
chr1_+_113929600
Show fit
|
3.04 |
ENST00000369558.5
ENST00000369561.8
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr9_-_128128102
Show fit
|
2.86 |
ENST00000617202.4
|
PTGES2
|
prostaglandin E synthase 2
|
|
chr6_-_31684040
Show fit
|
2.83 |
ENST00000375863.7
|
LY6G5C
|
lymphocyte antigen 6 family member G5C
|
|
chr11_-_65121780
Show fit
|
2.70 |
ENST00000525297.5
ENST00000529259.1
|
FAU
|
FAU ubiquitin like and ribosomal protein S30 fusion
|
|
chr1_+_153990749
Show fit
|
2.62 |
ENST00000651669.1
|
RPS27
|
ribosomal protein S27
|
|
chr13_+_102656933
Show fit
|
2.57 |
ENST00000650757.1
|
TPP2
|
tripeptidyl peptidase 2
|
|
chr8_+_32548210
Show fit
|
2.56 |
ENST00000523079.5
ENST00000650919.1
|
NRG1
|
neuregulin 1
|
|
chr19_+_4247074
Show fit
|
2.40 |
ENST00000262962.12
|
YJU2
|
YJU2 splicing factor homolog
|
|
chr6_+_85449584
Show fit
|
2.36 |
ENST00000369651.7
|
NT5E
|
5'-nucleotidase ecto
|
|
chr12_+_8123837
Show fit
|
2.32 |
ENST00000345999.9
ENST00000352620.9
ENST00000360500.5
|
CLEC4A
|
C-type lectin domain family 4 member A
|
|
chrX_+_108045050
Show fit
|
2.25 |
ENST00000458383.1
ENST00000217957.10
|
VSIG1
|
V-set and immunoglobulin domain containing 1
|
|
chr1_+_100345018
Show fit
|
2.24 |
ENST00000635056.2
ENST00000647005.1
|
CDC14A
|
cell division cycle 14A
|
|
chr1_+_40691689
Show fit
|
2.23 |
ENST00000427410.6
ENST00000447388.7
ENST00000425457.6
ENST00000453631.5
ENST00000456393.6
|
NFYC
|
nuclear transcription factor Y subunit gamma
|
|
chr9_+_75890664
Show fit
|
2.20 |
ENST00000376767.7
ENST00000674117.1
ENST00000376752.8
|
PCSK5
|
proprotein convertase subtilisin/kexin type 5
|
|
chr14_-_106360320
Show fit
|
2.19 |
ENST00000390615.2
|
IGHV3-33
|
immunoglobulin heavy variable 3-33
|
|
chr10_-_11532275
Show fit
|
2.15 |
ENST00000277575.5
|
USP6NL
|
USP6 N-terminal like
|
|
chr2_-_33599269
Show fit
|
2.13 |
ENST00000431950.1
ENST00000403368.1
ENST00000238823.13
|
FAM98A
|
family with sequence similarity 98 member A
|
|
chr7_+_93906557
Show fit
|
2.09 |
ENST00000248572.10
ENST00000429473.1
ENST00000430875.1
ENST00000428834.1
|
GNGT1
|
G protein subunit gamma transducin 1
|
|
chr22_+_31081310
Show fit
|
2.06 |
ENST00000426927.5
ENST00000482444.5
ENST00000440425.5
ENST00000333137.12
ENST00000358743.5
ENST00000347557.6
|
SMTN
|
smoothelin
|
|
chr4_+_69096494
Show fit
|
2.02 |
ENST00000508661.5
ENST00000622664.1
|
UGT2B7
|
UDP glucuronosyltransferase family 2 member B7
|
|
chr15_-_65517244
Show fit
|
1.97 |
ENST00000341861.9
|
DPP8
|
dipeptidyl peptidase 8
|
|
chr20_-_7940444
Show fit
|
1.97 |
ENST00000378789.4
|
HAO1
|
hydroxyacid oxidase 1
|
|
chr12_+_112906777
Show fit
|
1.93 |
ENST00000452357.7
ENST00000445409.7
|
OAS1
|
2'-5'-oligoadenylate synthetase 1
|
|
chrX_+_130401962
Show fit
|
1.91 |
ENST00000305536.11
ENST00000370947.1
|
RBMX2
|
RNA binding motif protein X-linked 2
|
|
chr12_-_10810168
Show fit
|
1.83 |
ENST00000240691.4
|
TAS2R9
|
taste 2 receptor member 9
|
|
chr12_-_10884244
Show fit
|
1.82 |
ENST00000543626.4
|
PRH1
|
proline rich protein HaeIII subfamily 1
|
|
chr12_+_75391078
Show fit
|
1.82 |
ENST00000550916.6
ENST00000378692.7
ENST00000320460.8
ENST00000547164.1
|
GLIPR1L2
|
GLIPR1 like 2
|
|
chr5_-_150700910
Show fit
|
1.81 |
ENST00000521464.1
ENST00000518917.5
ENST00000447771.6
ENST00000199814.9
|
RBM22
|
RNA binding motif protein 22
|
|
chrX_+_12906639
Show fit
|
1.77 |
ENST00000311912.5
|
TLR8
|
toll like receptor 8
|
|
chr4_+_69096467
Show fit
|
1.70 |
ENST00000305231.12
|
UGT2B7
|
UDP glucuronosyltransferase family 2 member B7
|
|
chrX_+_108044967
Show fit
|
1.69 |
ENST00000415430.7
|
VSIG1
|
V-set and immunoglobulin domain containing 1
|
|
chr4_+_15703057
Show fit
|
1.64 |
ENST00000265016.9
ENST00000382346.7
|
BST1
|
bone marrow stromal cell antigen 1
|
|
chr6_-_130956371
Show fit
|
1.59 |
ENST00000639623.1
ENST00000525193.5
ENST00000527659.5
|
EPB41L2
|
erythrocyte membrane protein band 4.1 like 2
|
|
chr9_+_75890639
Show fit
|
1.56 |
ENST00000545128.5
|
PCSK5
|
proprotein convertase subtilisin/kexin type 5
|
|
chr15_+_67125707
Show fit
|
1.51 |
ENST00000540846.6
|
SMAD3
|
SMAD family member 3
|
|
chr6_-_26285526
Show fit
|
1.50 |
ENST00000377727.2
|
H4C8
|
H4 clustered histone 8
|
|
chr19_+_9247344
Show fit
|
1.35 |
ENST00000641946.1
|
OR7E24
|
olfactory receptor family 7 subfamily E member 24
|
|
chr11_-_22625804
Show fit
|
1.34 |
ENST00000327470.6
|
FANCF
|
FA complementation group F
|
|
chr21_+_38256698
Show fit
|
1.28 |
ENST00000613499.4
ENST00000612702.4
ENST00000398925.5
ENST00000398928.5
ENST00000328656.8
ENST00000443341.5
|
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15
|
|
chr14_+_23988914
Show fit
|
1.27 |
ENST00000335125.11
|
DHRS4L2
|
dehydrogenase/reductase 4 like 2
|
|
chr19_+_49487510
Show fit
|
1.26 |
ENST00000679106.1
ENST00000621674.4
ENST00000391857.9
ENST00000678510.1
ENST00000467825.2
|
RPL13A
|
ribosomal protein L13a
|
|
chr19_+_852295
Show fit
|
1.16 |
ENST00000263621.2
|
ELANE
|
elastase, neutrophil expressed
|
|
chr1_+_78620432
Show fit
|
1.16 |
ENST00000370751.10
ENST00000459784.6
ENST00000680110.1
ENST00000680295.1
|
IFI44L
|
interferon induced protein 44 like
|
|
chr2_-_213151590
Show fit
|
1.09 |
ENST00000374319.8
ENST00000457361.5
ENST00000451136.6
ENST00000434687.6
|
IKZF2
|
IKAROS family zinc finger 2
|
|
chr5_+_55024250
Show fit
|
1.09 |
ENST00000231009.3
|
GZMK
|
granzyme K
|
|
chr1_+_160796157
Show fit
|
1.04 |
ENST00000263285.11
ENST00000368039.2
|
LY9
|
lymphocyte antigen 9
|
|
chr5_-_33984681
Show fit
|
1.01 |
ENST00000296589.9
|
SLC45A2
|
solute carrier family 45 member 2
|
|
chr5_-_78985288
Show fit
|
1.00 |
ENST00000264914.10
|
ARSB
|
arylsulfatase B
|
|
chr15_-_68229658
Show fit
|
1.00 |
ENST00000565471.6
ENST00000637494.1
ENST00000636314.1
ENST00000637667.1
ENST00000564752.1
ENST00000566347.5
ENST00000249806.11
ENST00000562767.2
|
CLN6
ENSG00000260007.3
|
CLN6 transmembrane ER protein
novel protein
|
|
chr6_-_26216673
Show fit
|
0.97 |
ENST00000541790.3
|
H2BC8
|
H2B clustered histone 8
|
|
chr5_-_100903252
Show fit
|
0.90 |
ENST00000231461.10
|
ST8SIA4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4
|
|
chr11_-_71448406
Show fit
|
0.85 |
ENST00000682708.1
ENST00000683287.1
ENST00000683714.1
ENST00000682880.1
|
DHCR7
|
7-dehydrocholesterol reductase
|
|
chr19_-_54313074
Show fit
|
0.83 |
ENST00000486742.2
ENST00000432233.8
|
LILRA5
|
leukocyte immunoglobulin like receptor A5
|
|
chr4_-_68951763
Show fit
|
0.83 |
ENST00000251566.9
|
UGT2A3
|
UDP glucuronosyltransferase family 2 member A3
|
|
chr4_+_26319636
Show fit
|
0.69 |
ENST00000342295.6
ENST00000506956.5
ENST00000512671.6
ENST00000345843.8
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region
|
|
chr8_+_2045037
Show fit
|
0.65 |
ENST00000262113.9
|
MYOM2
|
myomesin 2
|
|
chr17_-_19745602
Show fit
|
0.63 |
ENST00000444455.5
ENST00000439102.6
|
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1
|
|
chr18_+_63887698
Show fit
|
0.55 |
ENST00000457692.5
ENST00000299502.9
ENST00000413956.5
|
SERPINB2
|
serpin family B member 2
|
|
chr4_-_167234426
Show fit
|
0.53 |
ENST00000541354.5
ENST00000509854.5
ENST00000512681.5
ENST00000357545.9
ENST00000510741.5
ENST00000510403.5
|
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3
|
|
chr12_-_10453330
Show fit
|
0.49 |
ENST00000347831.9
ENST00000359151.8
|
KLRC1
|
killer cell lectin like receptor C1
|
|
chr6_-_127459364
Show fit
|
0.39 |
ENST00000487331.2
ENST00000483725.8
|
KIAA0408
|
KIAA0408
|
|
chr19_-_57457140
Show fit
|
0.31 |
ENST00000321039.5
|
VN1R1
|
vomeronasal 1 receptor 1
|
|
chr16_+_33827140
Show fit
|
0.25 |
ENST00000562905.2
|
IGHV3OR16-13
|
immunoglobulin heavy variable 3/OR16-13 (non-functional)
|
|
chr1_+_78620722
Show fit
|
0.19 |
ENST00000679848.1
|
IFI44L
|
interferon induced protein 44 like
|
|
chr20_-_44960348
Show fit
|
0.14 |
ENST00000372813.4
|
TOMM34
|
translocase of outer mitochondrial membrane 34
|
|
chr6_-_26271815
Show fit
|
0.11 |
ENST00000614378.1
|
H3C8
|
H3 clustered histone 8
|
|
chr1_+_150926336
Show fit
|
0.11 |
ENST00000271640.9
ENST00000448029.5
ENST00000368962.6
ENST00000534805.5
ENST00000368969.8
ENST00000368963.5
ENST00000498193.5
|
SETDB1
|
SET domain bifurcated histone lysine methyltransferase 1
|
|
chr2_+_156473569
Show fit
|
0.00 |
ENST00000409674.5
ENST00000409125.8
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2
|