Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
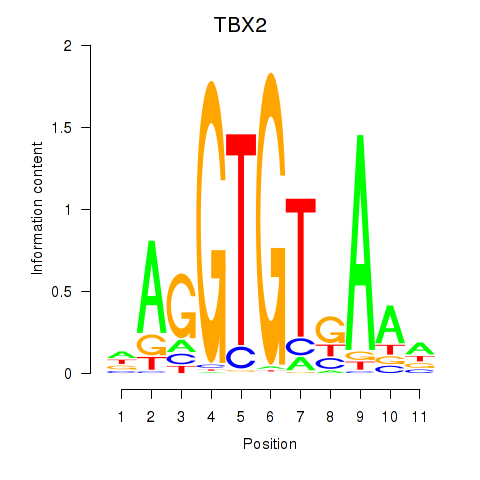
Results for TBX2
Z-value: 0.70
Transcription factors associated with TBX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX2
|
ENSG00000121068.14 | TBX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX2 | hg38_v1_chr17_+_61399835_61399851 | -0.07 | 3.3e-01 | Click! |
Activity profile of TBX2 motif
Sorted Z-values of TBX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 40.7 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 2.4 | 7.3 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 2.4 | 7.3 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 2.4 | 7.1 | GO:0009996 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) negative regulation of cell fate specification(GO:0009996) |
| 2.1 | 10.7 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 2.1 | 8.4 | GO:0090096 | metanephric cap development(GO:0072185) metanephric cap morphogenesis(GO:0072186) metanephric cap mesenchymal cell proliferation involved in metanephros development(GO:0090094) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 1.9 | 5.7 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 1.9 | 5.6 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) rRNA (guanine-N7)-methylation(GO:0070476) |
| 1.9 | 5.6 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 1.8 | 7.4 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 1.6 | 8.0 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 1.5 | 4.4 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 1.2 | 3.7 | GO:0051097 | negative regulation of helicase activity(GO:0051097) oligodendrocyte apoptotic process(GO:0097252) |
| 1.2 | 7.3 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 1.2 | 4.9 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 1.2 | 7.3 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 1.2 | 12.9 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 1.1 | 3.4 | GO:0071338 | male genitalia morphogenesis(GO:0048808) bronchiole development(GO:0060435) submandibular salivary gland formation(GO:0060661) positive regulation of fat cell proliferation(GO:0070346) hair follicle cell proliferation(GO:0071335) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) male anatomical structure morphogenesis(GO:0090598) |
| 1.1 | 3.3 | GO:0072183 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 1.0 | 3.1 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 1.0 | 7.9 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 1.0 | 3.8 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.9 | 2.8 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 0.9 | 2.8 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.9 | 5.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.9 | 2.6 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.9 | 3.4 | GO:0002357 | defense response to tumor cell(GO:0002357) |
| 0.8 | 2.4 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.8 | 3.1 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.8 | 7.8 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.8 | 2.3 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.8 | 3.0 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.7 | 2.9 | GO:0046223 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.7 | 2.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.7 | 2.0 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.7 | 4.7 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.7 | 3.9 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.6 | 8.3 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.6 | 5.7 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.6 | 3.1 | GO:0001315 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.6 | 3.7 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.6 | 3.0 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.5 | 5.4 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.5 | 9.9 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.5 | 3.1 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.5 | 1.5 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.5 | 3.0 | GO:0051958 | methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.5 | 2.0 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.5 | 2.5 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.5 | 2.0 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.5 | 1.9 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.5 | 7.7 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.4 | 2.2 | GO:2001107 | negative regulation of establishment of T cell polarity(GO:1903904) negative regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001107) |
| 0.4 | 1.8 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.4 | 3.5 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.4 | 1.7 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.4 | 1.7 | GO:1902617 | response to fluoride(GO:1902617) |
| 0.4 | 1.6 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.4 | 2.8 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.4 | 2.0 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.4 | 1.2 | GO:0042704 | uterine wall breakdown(GO:0042704) |
| 0.4 | 1.9 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.4 | 2.3 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.4 | 9.8 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.4 | 4.1 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.4 | 2.6 | GO:0060844 | arterial endothelial cell fate commitment(GO:0060844) blood vessel lumenization(GO:0072554) positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.4 | 2.2 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.4 | 14.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.3 | 3.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.3 | 1.7 | GO:0061083 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.3 | 8.8 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.3 | 1.7 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.3 | 2.6 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.3 | 1.3 | GO:0046778 | modification by virus of host mRNA processing(GO:0046778) |
| 0.3 | 3.5 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.3 | 1.9 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.3 | 2.8 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.3 | 3.6 | GO:0002860 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.3 | 2.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.3 | 0.6 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 0.3 | 5.7 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.3 | 1.7 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.3 | 4.4 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.3 | 3.0 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.3 | 4.0 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.3 | 1.1 | GO:0051582 | peptidyl-cysteine methylation(GO:0018125) positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.3 | 2.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.3 | 2.1 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.3 | 6.1 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.3 | 1.3 | GO:0043126 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.3 | 4.6 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.3 | 1.8 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.2 | 0.7 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.2 | 3.7 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.2 | 1.6 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 2.7 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.2 | 2.4 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.2 | 2.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 2.5 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.2 | 3.3 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.2 | 1.4 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.2 | 3.4 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.2 | 1.2 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.2 | 3.1 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.2 | 2.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.2 | 0.6 | GO:0099541 | trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.2 | 2.4 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.2 | 0.7 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.2 | 4.2 | GO:0007567 | parturition(GO:0007567) |
| 0.2 | 4.8 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.2 | 1.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.2 | 0.5 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.2 | 6.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 2.1 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 1.7 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 3.8 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.1 | 1.0 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 3.0 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.6 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.7 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.1 | 9.0 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 1.3 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 0.4 | GO:1903173 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.1 | 2.8 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.1 | 2.5 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 1.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 1.9 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 1.1 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 1.1 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.1 | 0.7 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.1 | 1.1 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 1.5 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.1 | 1.0 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.3 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.1 | 3.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 2.9 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.1 | 0.5 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 4.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 3.4 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 1.8 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 1.4 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.1 | 0.9 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 1.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 1.2 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 1.2 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.1 | 0.8 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.1 | 0.5 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) negative regulation of osteoclast development(GO:2001205) |
| 0.1 | 0.9 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.1 | 0.4 | GO:0032202 | telomere assembly(GO:0032202) scaRNA localization to Cajal body(GO:0090666) |
| 0.1 | 3.7 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.7 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 2.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 1.1 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 0.5 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 0.6 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.1 | 3.5 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 0.2 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 | 5.4 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.8 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.6 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 0.5 | GO:0033630 | positive regulation of cell adhesion mediated by integrin(GO:0033630) |
| 0.0 | 0.5 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.3 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.6 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 1.6 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 2.3 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 | 0.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 1.1 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.1 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.0 | 1.9 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 1.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 7.1 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.0 | 0.2 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.1 | GO:0015888 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.0 | 3.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.1 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 1.5 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 5.8 | GO:0007051 | spindle organization(GO:0007051) |
| 0.0 | 0.7 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 2.8 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 0.8 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.4 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.4 | GO:0021756 | striatum development(GO:0021756) |
| 0.0 | 0.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.5 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.2 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.8 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.9 | GO:0055010 | ventricular cardiac muscle tissue morphogenesis(GO:0055010) |
| 0.0 | 1.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.2 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 1.6 | GO:0001523 | retinoid metabolic process(GO:0001523) |
| 0.0 | 2.1 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 1.3 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 1.6 | GO:0051321 | meiotic cell cycle(GO:0051321) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 40.7 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 1.7 | 11.7 | GO:0034715 | U7 snRNP(GO:0005683) pICln-Sm protein complex(GO:0034715) |
| 1.3 | 11.9 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 1.1 | 3.4 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 1.0 | 3.8 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.9 | 8.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.8 | 3.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.7 | 7.9 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.7 | 10.1 | GO:0070938 | contractile ring(GO:0070938) |
| 0.7 | 8.7 | GO:0030677 | ribonuclease P complex(GO:0030677) |
| 0.7 | 5.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.6 | 12.9 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.6 | 7.3 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.6 | 6.6 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.6 | 1.7 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.6 | 1.7 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.5 | 1.5 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.5 | 1.9 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.5 | 4.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.4 | 2.6 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.4 | 7.4 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.4 | 9.8 | GO:0032982 | myosin filament(GO:0032982) |
| 0.4 | 5.7 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.3 | 3.4 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.3 | 1.7 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.3 | 2.0 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.3 | 2.6 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.3 | 2.0 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.3 | 5.7 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.3 | 3.7 | GO:0090543 | Flemming body(GO:0090543) |
| 0.3 | 1.3 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.3 | 1.0 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.2 | 2.0 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) ERCC4-ERCC1 complex(GO:0070522) |
| 0.2 | 14.7 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.2 | 1.1 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.2 | 1.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.2 | 2.2 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.2 | 2.8 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.2 | 3.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 3.9 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.2 | 3.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 2.5 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.2 | 2.8 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.2 | 4.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 9.8 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 4.0 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 6.4 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.2 | 1.5 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.2 | 2.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.2 | 3.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.2 | 5.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.2 | 0.8 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.2 | 2.1 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.1 | 1.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 2.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.7 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 4.0 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 3.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 1.7 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 4.1 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 2.1 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 0.7 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 2.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 5.6 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 1.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 4.0 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 1.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.6 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.5 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 0.3 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 4.3 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 1.7 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 3.0 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 3.0 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 5.9 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.1 | 6.4 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.1 | 4.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.4 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.1 | 2.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 3.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 6.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 2.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 1.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.5 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 1.3 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.7 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 1.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 2.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.2 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 1.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 9.6 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 1.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 1.9 | GO:0097525 | spliceosomal snRNP complex(GO:0097525) |
| 0.0 | 2.8 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 1.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 2.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 2.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 21.0 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.3 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 2.3 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 1.8 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.0 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 1.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.1 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.4 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 1.3 | 5.2 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 1.1 | 5.7 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 1.0 | 3.8 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.9 | 5.6 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.9 | 7.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.9 | 3.4 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.8 | 2.4 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.7 | 2.8 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.7 | 2.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.6 | 3.8 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.6 | 3.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.6 | 6.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.6 | 4.6 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.5 | 3.0 | GO:0008518 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.5 | 2.7 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.4 | 8.8 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.4 | 3.5 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.4 | 2.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.4 | 12.9 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.4 | 3.7 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.4 | 6.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.4 | 5.4 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.4 | 2.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.4 | 7.9 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.3 | 1.0 | GO:0032428 | sphingolipid activator protein activity(GO:0030290) beta-N-acetylgalactosaminidase activity(GO:0032428) |
| 0.3 | 8.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.3 | 1.3 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.3 | 3.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.3 | 3.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.3 | 2.1 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.3 | 2.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.3 | 1.2 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.3 | 2.0 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.3 | 5.7 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.3 | 6.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 1.9 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.3 | 3.0 | GO:0046790 | virion binding(GO:0046790) |
| 0.3 | 3.8 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.3 | 3.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.3 | 7.3 | GO:0071949 | FAD binding(GO:0071949) |
| 0.2 | 1.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 2.0 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.2 | 1.2 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.2 | 7.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 1.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.2 | 4.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 2.8 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.2 | 0.7 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.2 | 0.8 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.2 | 2.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 3.0 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 0.5 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.2 | 2.6 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.2 | 0.5 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.2 | 1.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 3.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 1.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.4 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 0.7 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.1 | 1.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 2.0 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.7 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.1 | 2.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 2.8 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 3.6 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 1.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 2.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 1.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 2.8 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 4.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 1.1 | GO:0030883 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.1 | 3.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 3.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 0.8 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.1 | 1.1 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.1 | 2.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 1.3 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 3.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.4 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.1 | 3.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 8.3 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 4.5 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.1 | 20.5 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.1 | 0.6 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.1 | 0.3 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 6.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 1.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 1.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 1.8 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.8 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.6 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.6 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 7.9 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 1.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 3.5 | GO:0061650 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.1 | 2.1 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 4.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 4.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 2.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 2.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 2.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 1.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 0.3 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 2.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 11.8 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 2.0 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.1 | 0.8 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 7.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.9 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 2.3 | GO:0008137 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.1 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 8.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 11.9 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.1 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 1.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.3 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 1.6 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 1.9 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 2.3 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.5 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 4.4 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 8.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.8 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.4 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.4 | 6.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.4 | 11.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.4 | 26.8 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.3 | 7.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.2 | 3.3 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 8.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 11.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 5.0 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 5.7 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 2.0 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 2.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 3.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 5.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 7.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 2.8 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 5.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 5.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.0 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 4.7 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 3.5 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 4.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 2.3 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 2.8 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 2.8 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 1.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 7.2 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 4.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.4 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 2.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.8 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.8 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 1.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.1 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 1.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.1 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.1 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 9.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.5 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 10.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.7 | 10.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.6 | 8.9 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.6 | 9.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.4 | 14.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.4 | 11.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.4 | 10.6 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.3 | 5.7 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.3 | 9.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.3 | 3.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.2 | 4.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 2.8 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 2.0 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.2 | 9.0 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.2 | 5.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 8.0 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 10.1 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.2 | 28.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 8.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.8 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 2.6 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 2.0 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.1 | 3.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 5.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 2.8 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 3.8 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 2.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 4.2 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 2.9 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 2.5 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 3.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.7 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 1.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 4.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 0.6 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 2.0 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 2.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 1.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 3.8 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.1 | 1.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 0.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 3.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 3.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 5.3 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 7.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 4.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.6 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.9 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.0 | 2.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 2.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 2.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.4 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 1.6 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 2.3 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.1 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |