Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
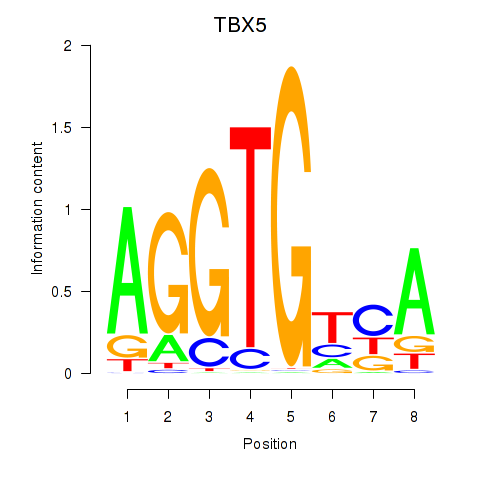
Results for TBX5
Z-value: 1.06
Transcription factors associated with TBX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX5
|
ENSG00000089225.20 | TBX5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX5 | hg38_v1_chr12_-_114406133_114406150 | 0.46 | 9.1e-13 | Click! |
Activity profile of TBX5 motif
Sorted Z-values of TBX5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 14.7 | GO:1904397 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 4.5 | 36.1 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 4.3 | 25.7 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 4.1 | 16.3 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 4.0 | 12.1 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 3.7 | 11.0 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 3.6 | 32.7 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 3.3 | 9.8 | GO:0060032 | notochord regression(GO:0060032) |
| 2.9 | 8.7 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 2.6 | 7.8 | GO:1903937 | response to acrylamide(GO:1903937) |
| 2.6 | 7.7 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 2.3 | 9.0 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 2.1 | 8.5 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 2.1 | 8.4 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 2.1 | 10.4 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 2.1 | 6.2 | GO:0009720 | detection of hormone stimulus(GO:0009720) |
| 2.1 | 6.2 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 1.9 | 7.4 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 1.8 | 5.5 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 1.7 | 10.4 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 1.7 | 5.0 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 1.5 | 4.6 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 1.5 | 17.6 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 1.4 | 4.2 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 1.4 | 6.9 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 1.4 | 9.6 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 1.3 | 1.3 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 1.3 | 3.9 | GO:0060005 | vestibular reflex(GO:0060005) |
| 1.3 | 5.1 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 1.3 | 3.8 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 1.3 | 3.8 | GO:0099557 | trans-synaptic signaling by trans-synaptic complex, modulating synaptic transmission(GO:0099557) |
| 1.2 | 3.7 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 1.2 | 6.2 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 1.2 | 3.7 | GO:0021718 | pons maturation(GO:0021586) superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 1.2 | 4.8 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 1.2 | 4.6 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 1.1 | 3.4 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 1.1 | 4.5 | GO:0032904 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 1.1 | 6.6 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 1.1 | 13.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 1.1 | 5.4 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 1.0 | 2.9 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 1.0 | 3.9 | GO:0035283 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 1.0 | 14.3 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 1.0 | 7.6 | GO:1903960 | negative regulation of anion transmembrane transport(GO:1903960) |
| 0.9 | 6.6 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.9 | 3.7 | GO:2000196 | positive regulation of female gonad development(GO:2000196) |
| 0.9 | 20.3 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.9 | 9.9 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.9 | 5.3 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.9 | 7.0 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.9 | 3.4 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.8 | 0.8 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.8 | 6.6 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.8 | 2.4 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.8 | 4.0 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) |
| 0.8 | 5.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.7 | 3.6 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.7 | 3.5 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.7 | 6.2 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.7 | 2.0 | GO:0061591 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.6 | 1.3 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.6 | 8.0 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.6 | 1.1 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.5 | 9.5 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.5 | 2.0 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.5 | 1.9 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.4 | 6.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.4 | 3.6 | GO:2000332 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.4 | 2.7 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.4 | 2.2 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.4 | 2.6 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.4 | 3.9 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.4 | 2.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.4 | 17.6 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.4 | 3.3 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 0.4 | 9.9 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.4 | 14.6 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.4 | 2.0 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.4 | 6.4 | GO:0044126 | growth of symbiont in host(GO:0044117) regulation of growth of symbiont in host(GO:0044126) |
| 0.4 | 3.9 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.4 | 1.2 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) sarcomerogenesis(GO:0048769) |
| 0.4 | 8.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.4 | 1.9 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.4 | 6.8 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.4 | 2.6 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.4 | 1.5 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.4 | 10.1 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.4 | 7.4 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.4 | 1.9 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.4 | 1.1 | GO:0001207 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.4 | 5.1 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.3 | 15.0 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.3 | 2.4 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.3 | 1.0 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.3 | 2.7 | GO:0046137 | negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.3 | 15.0 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.3 | 3.9 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.3 | 2.6 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.3 | 6.4 | GO:0035646 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.3 | 8.8 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 1.2 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.3 | 0.9 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.3 | 10.8 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.3 | 7.5 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.3 | 2.4 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.3 | 4.5 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.3 | 2.0 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.3 | 0.8 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.3 | 2.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.3 | 1.4 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.3 | 0.5 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.3 | 6.0 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.3 | 4.6 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.3 | 4.0 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.3 | 1.0 | GO:0010193 | response to ozone(GO:0010193) |
| 0.2 | 3.0 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.2 | 1.5 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.2 | 2.2 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.2 | 2.9 | GO:0051775 | response to redox state(GO:0051775) |
| 0.2 | 2.1 | GO:1901626 | regulation of postsynaptic membrane organization(GO:1901626) |
| 0.2 | 1.7 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.2 | 6.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 1.1 | GO:0002554 | serotonin production involved in inflammatory response(GO:0002351) serotonin secretion involved in inflammatory response(GO:0002442) serotonin secretion by platelet(GO:0002554) |
| 0.2 | 19.0 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.2 | 4.0 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.2 | 21.2 | GO:0007422 | peripheral nervous system development(GO:0007422) |
| 0.2 | 1.4 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.2 | 4.8 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.2 | 14.3 | GO:0007602 | phototransduction(GO:0007602) |
| 0.2 | 2.6 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.2 | 3.5 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.2 | 1.9 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.2 | 2.2 | GO:0097070 | ductus arteriosus closure(GO:0097070) |
| 0.2 | 1.4 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.2 | 3.9 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.2 | 2.5 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.2 | 7.0 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.2 | 2.4 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 2.7 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.1 | 2.5 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 0.4 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 3.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.3 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.1 | 3.4 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 4.0 | GO:0045187 | regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.1 | 2.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 10.1 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.1 | 1.9 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.1 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.1 | 1.0 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.1 | 0.5 | GO:0090240 | positive regulation of histone H4 acetylation(GO:0090240) |
| 0.1 | 2.6 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 1.6 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 5.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 0.5 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.1 | 5.2 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.1 | 2.0 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 1.9 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.1 | 1.7 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 1.1 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 1.2 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 10.7 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.1 | 0.4 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 2.3 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 5.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 36.9 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.1 | 4.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.7 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.1 | 9.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 2.8 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.1 | 2.8 | GO:0048536 | spleen development(GO:0048536) |
| 0.1 | 1.1 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 1.8 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.1 | 1.0 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 | 4.0 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.1 | 0.2 | GO:2000697 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 0.1 | 0.8 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.1 | 0.4 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.1 | 4.5 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 2.9 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 2.5 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 5.1 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.1 | 6.5 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.1 | 3.5 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.1 | 2.4 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.1 | 4.4 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.1 | 2.0 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 2.7 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 3.9 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 1.1 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 4.6 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 2.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 1.8 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 4.0 | GO:1901799 | negative regulation of proteasomal protein catabolic process(GO:1901799) |
| 0.0 | 2.4 | GO:0007270 | neuron-neuron synaptic transmission(GO:0007270) |
| 0.0 | 2.2 | GO:0007188 | adenylate cyclase-modulating G-protein coupled receptor signaling pathway(GO:0007188) |
| 0.0 | 0.8 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.3 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 2.8 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 1.2 | GO:0048599 | oocyte development(GO:0048599) |
| 0.0 | 1.0 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 15.2 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.6 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 3.8 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 3.0 | GO:0035113 | embryonic limb morphogenesis(GO:0030326) embryonic appendage morphogenesis(GO:0035113) |
| 0.0 | 1.9 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.2 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.0 | 1.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.9 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 1.8 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.5 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.0 | 2.3 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 1.1 | GO:0016079 | synaptic vesicle exocytosis(GO:0016079) |
| 0.0 | 2.1 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.6 | GO:0009953 | dorsal/ventral pattern formation(GO:0009953) |
| 0.0 | 1.2 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 1.1 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 32.7 | GO:0035976 | AP1 complex(GO:0035976) |
| 2.1 | 22.9 | GO:0005955 | calcineurin complex(GO:0005955) |
| 1.9 | 7.8 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 1.9 | 7.5 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 1.3 | 10.4 | GO:0045179 | apical cortex(GO:0045179) |
| 1.3 | 3.9 | GO:0032426 | stereocilium tip(GO:0032426) |
| 1.2 | 6.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 1.2 | 11.0 | GO:0072687 | meiotic spindle(GO:0072687) |
| 1.2 | 4.7 | GO:0070695 | FHF complex(GO:0070695) |
| 1.1 | 17.5 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 1.1 | 4.3 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 0.8 | 4.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.7 | 3.7 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.7 | 3.7 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.7 | 2.2 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.6 | 1.3 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.6 | 3.7 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.6 | 4.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.6 | 8.6 | GO:0097433 | dense body(GO:0097433) |
| 0.5 | 1.6 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.5 | 6.9 | GO:0060077 | inhibitory synapse(GO:0060077) clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.5 | 5.0 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.5 | 12.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.5 | 32.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.4 | 6.4 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.4 | 2.9 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.4 | 2.0 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.4 | 17.8 | GO:0097546 | ciliary base(GO:0097546) |
| 0.4 | 1.9 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.3 | 7.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.3 | 8.5 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.3 | 5.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.3 | 11.5 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.3 | 0.8 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.3 | 3.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.2 | 2.2 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.2 | 6.8 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.2 | 34.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.2 | 2.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.2 | 3.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 4.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 1.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.2 | 6.0 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.2 | 2.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.2 | 6.2 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 29.6 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.2 | 21.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.2 | 1.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 3.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 1.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 12.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 2.8 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 8.0 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 8.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 6.2 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 3.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 4.6 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 4.8 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.1 | 2.6 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 2.0 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 25.4 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.1 | 1.8 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.1 | 4.8 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 0.5 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 3.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 1.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 0.5 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 1.7 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 5.6 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.1 | 3.6 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 1.0 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 11.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 1.1 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 5.3 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 3.7 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.1 | 2.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 2.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.9 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 11.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 5.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.8 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 1.1 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 1.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.9 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 2.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 2.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 10.0 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 2.5 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 1.7 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 7.4 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 3.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 5.5 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.5 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 5.2 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 1.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 22.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 4.3 | 25.7 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 4.0 | 36.1 | GO:0043426 | MRF binding(GO:0043426) |
| 3.7 | 14.7 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 3.7 | 11.0 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 2.3 | 9.0 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 2.0 | 25.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 1.8 | 16.3 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 1.8 | 5.4 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 1.7 | 8.7 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 1.7 | 5.1 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 1.7 | 5.0 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 1.6 | 6.5 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 1.6 | 9.6 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 1.5 | 6.2 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 1.4 | 6.8 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 1.3 | 34.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 1.2 | 3.7 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 1.1 | 4.5 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 1.1 | 3.4 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 1.1 | 3.4 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 1.1 | 6.4 | GO:0008142 | oxysterol binding(GO:0008142) |
| 1.0 | 5.0 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.9 | 3.7 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.9 | 3.6 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.9 | 2.6 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.9 | 6.9 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.9 | 3.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.8 | 6.6 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.8 | 4.0 | GO:0034648 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.7 | 3.0 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.7 | 4.3 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) glycerol transmembrane transporter activity(GO:0015168) |
| 0.7 | 2.1 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.7 | 8.4 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) TPR domain binding(GO:0030911) |
| 0.7 | 6.2 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.7 | 2.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.7 | 2.6 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) |
| 0.7 | 5.9 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.7 | 5.3 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.7 | 13.8 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.6 | 3.9 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.6 | 7.7 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.6 | 1.9 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.6 | 38.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.6 | 1.9 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.6 | 3.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.6 | 4.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.6 | 5.5 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.5 | 1.6 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.5 | 2.2 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.5 | 4.3 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.5 | 3.7 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.5 | 5.1 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.5 | 7.6 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.5 | 5.0 | GO:0015250 | water channel activity(GO:0015250) |
| 0.5 | 9.9 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.5 | 10.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.5 | 3.8 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.5 | 4.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.4 | 2.2 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.4 | 4.5 | GO:0005549 | odorant binding(GO:0005549) |
| 0.4 | 3.6 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.4 | 6.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.4 | 2.6 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.4 | 17.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.4 | 7.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.4 | 1.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.4 | 1.6 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.4 | 2.7 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.4 | 2.2 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.3 | 1.0 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.3 | 2.7 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.3 | 15.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.3 | 7.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 0.9 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.3 | 2.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.3 | 4.8 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.3 | 4.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.3 | 2.6 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.3 | 0.8 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.3 | 1.9 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.3 | 3.9 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.3 | 4.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.3 | 8.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.3 | 8.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 2.2 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.2 | 3.9 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) retinal binding(GO:0016918) |
| 0.2 | 4.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 9.8 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.2 | 2.8 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.2 | 1.9 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.2 | 3.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.2 | 3.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 3.4 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.2 | 2.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.2 | 3.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 1.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.2 | 1.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 5.4 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.2 | 18.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 0.5 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.2 | 2.9 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.2 | 4.7 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.2 | 0.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 1.0 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.2 | 2.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 2.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 8.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 52.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 1.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 4.5 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 2.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 6.1 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.1 | 3.5 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 7.5 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 2.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.4 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 2.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 2.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 1.1 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 2.5 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 6.1 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 0.4 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.1 | 11.3 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 1.1 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 1.0 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 5.8 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 1.7 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 14.0 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.1 | 2.8 | GO:0016248 | channel inhibitor activity(GO:0016248) |
| 0.1 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 14.3 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 0.7 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 1.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 21.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 0.2 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.1 | 1.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 1.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 1.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 2.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 3.8 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 2.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 3.8 | GO:0020037 | heme binding(GO:0020037) |
| 0.1 | 1.0 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 4.9 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.1 | 4.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 2.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.3 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 3.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 1.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 1.0 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 1.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.5 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 3.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.0 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 1.0 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 7.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 5.8 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 6.1 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 2.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 3.4 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 2.4 | GO:0005516 | calmodulin binding(GO:0005516) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 32.7 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.6 | 16.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.5 | 14.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.4 | 29.1 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.3 | 3.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.3 | 22.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.3 | 10.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.3 | 11.8 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.3 | 3.9 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.3 | 10.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 7.7 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.2 | 16.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 1.4 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.2 | 6.6 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.2 | 9.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.2 | 6.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.2 | 9.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.2 | 1.9 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.2 | 1.7 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.2 | 9.6 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 2.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 2.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 3.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 4.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 1.7 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 3.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 3.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 10.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 5.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 12.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 3.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 4.1 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 5.8 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 3.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 1.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.9 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 0.8 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 4.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 2.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 2.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.9 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.5 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 1.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 5.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.5 | PID BCR 5PATHWAY | BCR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 32.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.7 | 23.9 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.6 | 8.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.5 | 10.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.5 | 16.0 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.4 | 3.5 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.3 | 14.4 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.3 | 4.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 7.8 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.3 | 6.9 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.3 | 6.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.3 | 4.5 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.3 | 23.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.3 | 8.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.3 | 6.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.3 | 5.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.3 | 4.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.3 | 6.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.3 | 49.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.3 | 4.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 8.7 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.3 | 15.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.3 | 8.7 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.2 | 2.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.2 | 15.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.2 | 1.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 6.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 4.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.2 | 8.1 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.2 | 2.6 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.2 | 39.8 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.2 | 1.7 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 2.0 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.2 | 3.0 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 2.1 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 6.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 3.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 3.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 5.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 9.3 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 1.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 6.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 2.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 2.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 2.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 3.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 5.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 0.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 1.0 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.4 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 5.2 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 5.4 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 2.0 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 2.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 5.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.7 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 3.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.8 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 2.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 2.2 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 1.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |