Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for TCF3_MYOG
Z-value: 1.31
Transcription factors associated with TCF3_MYOG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
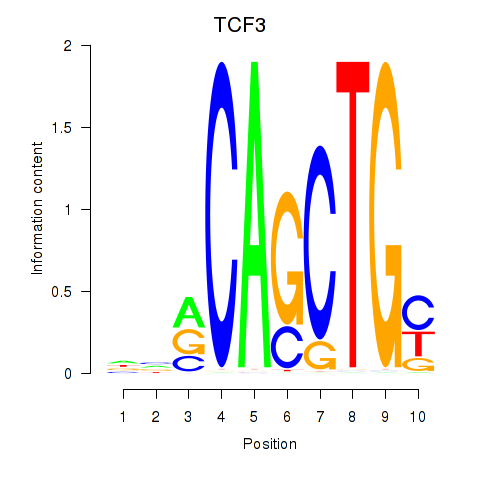
TCF3
|
ENSG00000071564.17 | TCF3 |
|
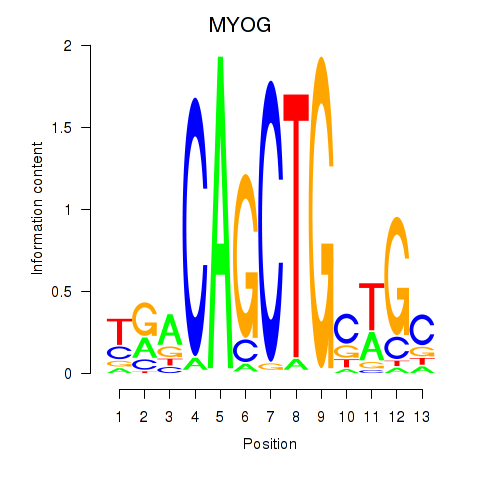
MYOG
|
ENSG00000122180.5 | MYOG |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MYOG | hg38_v1_chr1_-_203086001_203086019 | -0.34 | 2.4e-07 | Click! |
| TCF3 | hg38_v1_chr19_-_1652576_1652622 | -0.04 | 5.8e-01 | Click! |
Activity profile of TCF3_MYOG motif
Sorted Z-values of TCF3_MYOG motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TCF3_MYOG
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.7 | 38.8 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 6.3 | 25.0 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 5.0 | 14.9 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 4.5 | 13.4 | GO:0010768 | negative regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010768) transmission of virus(GO:0019089) dissemination or transmission of symbiont from host(GO:0044007) dissemination or transmission of organism from other organism involved in symbiotic interaction(GO:0051821) |
| 4.5 | 13.4 | GO:0008615 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 biosynthetic process(GO:0042819) |
| 4.0 | 11.9 | GO:0060739 | mesenchymal-epithelial cell signaling involved in prostate gland development(GO:0060739) |
| 3.9 | 15.6 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 3.6 | 10.9 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 3.6 | 17.9 | GO:1904636 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 3.4 | 10.3 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) regulation of embryonic cell shape(GO:0016476) |
| 3.3 | 9.9 | GO:0021592 | fourth ventricle development(GO:0021592) third ventricle development(GO:0021678) |
| 3.0 | 8.9 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 2.8 | 17.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 2.6 | 17.9 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 2.5 | 12.7 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 2.5 | 35.6 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 2.5 | 9.8 | GO:0018032 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 2.4 | 7.3 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 2.4 | 7.2 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) |
| 2.1 | 4.3 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 2.1 | 6.2 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 2.0 | 22.0 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 2.0 | 25.7 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 1.9 | 13.5 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 1.9 | 5.7 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 1.8 | 12.9 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 1.8 | 9.0 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 1.8 | 15.8 | GO:0031536 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) positive regulation of exit from mitosis(GO:0031536) |
| 1.7 | 10.5 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 1.7 | 5.0 | GO:0006113 | fermentation(GO:0006113) regulation of fermentation(GO:0043465) |
| 1.6 | 6.6 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 1.6 | 6.6 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 1.6 | 4.8 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 1.6 | 7.9 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 1.6 | 22.0 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 1.6 | 1.6 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 1.5 | 12.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 1.5 | 7.5 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 1.5 | 14.7 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 1.5 | 20.6 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) negative regulation of synapse assembly(GO:0051964) |
| 1.5 | 22.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 1.4 | 25.4 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 1.4 | 6.8 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 1.3 | 9.4 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 1.3 | 4.0 | GO:1901355 | response to rapamycin(GO:1901355) |
| 1.3 | 13.2 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.3 | 15.8 | GO:0009414 | response to water deprivation(GO:0009414) |
| 1.3 | 5.3 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 1.3 | 18.8 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 1.2 | 12.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 1.2 | 2.5 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 1.2 | 14.5 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 1.2 | 5.8 | GO:0042374 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 1.1 | 3.3 | GO:0097069 | response to human chorionic gonadotropin(GO:0044752) cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 1.1 | 3.3 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 1.1 | 16.3 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 1.1 | 2.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 1.1 | 5.3 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 1.1 | 5.3 | GO:0015862 | uridine transport(GO:0015862) |
| 1.1 | 7.4 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 1.0 | 4.1 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 1.0 | 76.0 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 1.0 | 17.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 1.0 | 70.9 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 1.0 | 16.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 1.0 | 4.8 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 1.0 | 5.8 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) glucose 1-phosphate metabolic process(GO:0019255) |
| 1.0 | 6.7 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 1.0 | 5.7 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.9 | 14.0 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.9 | 2.7 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.9 | 2.7 | GO:0072428 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.9 | 5.3 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.9 | 6.2 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.9 | 19.6 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.8 | 5.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.8 | 1.7 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.8 | 9.9 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.8 | 11.5 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.8 | 4.9 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.8 | 12.0 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.8 | 1.6 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.8 | 4.7 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.8 | 8.5 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.8 | 3.8 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.8 | 3.0 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.8 | 11.3 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.7 | 2.2 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.7 | 10.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.7 | 2.9 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.7 | 2.9 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.7 | 2.9 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.7 | 9.2 | GO:0009629 | response to gravity(GO:0009629) |
| 0.7 | 7.8 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.7 | 7.7 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.7 | 1.4 | GO:0018963 | phthalate metabolic process(GO:0018963) |
| 0.7 | 4.1 | GO:0003164 | His-Purkinje system development(GO:0003164) |
| 0.7 | 10.3 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.7 | 3.4 | GO:0007197 | regulation of vascular smooth muscle contraction(GO:0003056) adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.6 | 3.2 | GO:0061738 | late endosomal microautophagy(GO:0061738) negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.6 | 2.6 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.6 | 8.2 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.6 | 11.2 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.6 | 5.6 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.6 | 1.8 | GO:0002876 | positive regulation of chronic inflammatory response to antigenic stimulus(GO:0002876) |
| 0.6 | 8.6 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.6 | 3.1 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.6 | 3.0 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.6 | 3.6 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.6 | 7.2 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.6 | 2.4 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.6 | 1.8 | GO:0071314 | cellular response to cocaine(GO:0071314) response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.6 | 6.5 | GO:0015868 | purine nucleotide transport(GO:0015865) purine ribonucleotide transport(GO:0015868) |
| 0.6 | 3.5 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.6 | 1.7 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.6 | 1.7 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.6 | 4.0 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.6 | 19.0 | GO:0061641 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.6 | 5.0 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.6 | 6.7 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.6 | 5.0 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.5 | 6.6 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.5 | 2.7 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.5 | 5.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.5 | 2.6 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.5 | 3.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.5 | 4.7 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.5 | 2.1 | GO:0097350 | neutrophil clearance(GO:0097350) |
| 0.5 | 1.0 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.5 | 3.6 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.5 | 1.0 | GO:1903438 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.5 | 1.5 | GO:0031448 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.5 | 7.4 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.5 | 2.5 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.5 | 4.9 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.5 | 1.5 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.5 | 3.8 | GO:0009597 | detection of virus(GO:0009597) |
| 0.5 | 3.3 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.5 | 2.4 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.5 | 2.9 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.5 | 2.4 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.5 | 7.4 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.5 | 3.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.5 | 5.5 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.5 | 2.8 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.5 | 6.4 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.5 | 5.9 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.5 | 4.6 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.5 | 2.7 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.5 | 4.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.4 | 5.4 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.4 | 2.7 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.4 | 4.0 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.4 | 7.8 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.4 | 12.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.4 | 14.4 | GO:1904376 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.4 | 3.4 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.4 | 0.8 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.4 | 0.8 | GO:1905247 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.4 | 1.2 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.4 | 5.5 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.4 | 4.3 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.4 | 8.9 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.4 | 8.8 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.4 | 1.5 | GO:0061360 | optic nerve formation(GO:0021634) optic chiasma development(GO:0061360) regulation of optic nerve formation(GO:2000595) positive regulation of optic nerve formation(GO:2000597) |
| 0.4 | 3.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.4 | 21.7 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.4 | 4.6 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.4 | 7.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.4 | 6.0 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.4 | 1.1 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.4 | 0.7 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.4 | 6.3 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.4 | 34.9 | GO:0032392 | DNA geometric change(GO:0032392) |
| 0.4 | 1.8 | GO:0035747 | natural killer cell chemotaxis(GO:0035747) |
| 0.4 | 1.1 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.4 | 2.2 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.4 | 13.6 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.3 | 1.0 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.3 | 1.7 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.3 | 4.0 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.3 | 2.0 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.3 | 39.5 | GO:0031424 | keratinization(GO:0031424) |
| 0.3 | 8.5 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.3 | 1.9 | GO:0006388 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.3 | 1.9 | GO:0097319 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.3 | 13.4 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.3 | 2.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.3 | 35.9 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.3 | 6.6 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.3 | 0.3 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.3 | 2.8 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.3 | 4.0 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.3 | 19.0 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.3 | 3.7 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.3 | 0.6 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.3 | 7.0 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.3 | 1.5 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.3 | 11.5 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.3 | 3.6 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.3 | 2.6 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.3 | 0.9 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.3 | 2.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.3 | 23.4 | GO:0048207 | vesicle coating(GO:0006901) vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.3 | 5.6 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.3 | 1.4 | GO:0097490 | sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.3 | 8.6 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.3 | 3.1 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.3 | 4.1 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.3 | 1.9 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.3 | 1.6 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.3 | 2.1 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.3 | 0.8 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.3 | 2.7 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.3 | 0.8 | GO:1990009 | response to high light intensity(GO:0009644) cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) retinal cell apoptotic process(GO:1990009) |
| 0.3 | 3.9 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.3 | 1.3 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.3 | 2.5 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.2 | 1.5 | GO:0061743 | motor learning(GO:0061743) |
| 0.2 | 1.0 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.2 | 1.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.2 | 7.8 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.2 | 0.7 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.2 | 3.8 | GO:0042761 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.2 | 1.4 | GO:0009624 | response to nematode(GO:0009624) response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.2 | 0.9 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.2 | 4.3 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.2 | 2.5 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.2 | 0.7 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.2 | 2.9 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 | 0.9 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.2 | 1.5 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.2 | 6.4 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 1.7 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.2 | 7.7 | GO:0006378 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.2 | 0.4 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.2 | 1.0 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.2 | 1.4 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.2 | 2.0 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.2 | 1.8 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.2 | 1.2 | GO:0061767 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of lung blood pressure(GO:0061767) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.2 | 6.0 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.2 | 3.2 | GO:0042473 | outer ear morphogenesis(GO:0042473) |
| 0.2 | 4.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.2 | 1.1 | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) |
| 0.2 | 0.7 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.2 | 8.1 | GO:0048255 | mRNA stabilization(GO:0048255) |
| 0.2 | 8.3 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.2 | 0.4 | GO:0070244 | negative regulation of thymocyte apoptotic process(GO:0070244) |
| 0.2 | 2.9 | GO:0097205 | renal filtration(GO:0097205) |
| 0.2 | 2.6 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.2 | 16.5 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.2 | 0.7 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.2 | 7.0 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.2 | 0.7 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.2 | 0.4 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.2 | 0.4 | GO:0060437 | lung growth(GO:0060437) |
| 0.2 | 1.9 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.2 | 14.0 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.2 | 1.2 | GO:0060923 | regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003256) arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) positive regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061419) blood vessel lumenization(GO:0072554) blood vessel endothelial cell fate specification(GO:0097101) positive regulation of ephrin receptor signaling pathway(GO:1901189) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.2 | 2.1 | GO:0043586 | tongue development(GO:0043586) |
| 0.2 | 12.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.2 | 2.2 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.2 | 0.8 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.2 | 7.8 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.2 | 1.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.2 | 3.2 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.2 | 5.4 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.2 | 1.7 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 11.3 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 2.5 | GO:0021681 | cerebellar granular layer development(GO:0021681) |
| 0.1 | 10.2 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.1 | 3.0 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.1 | 3.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 1.5 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 | 0.4 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.1 | 0.1 | GO:0038129 | ERBB3 signaling pathway(GO:0038129) |
| 0.1 | 0.5 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.1 | 2.8 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 9.7 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.1 | 0.7 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.1 | 3.5 | GO:0072662 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 4.8 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 3.3 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 0.3 | GO:0071671 | regulation of smooth muscle cell chemotaxis(GO:0071671) positive regulation of smooth muscle cell chemotaxis(GO:0071673) |
| 0.1 | 2.0 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.1 | 0.6 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.1 | 1.3 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.1 | 1.0 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.1 | 0.2 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.1 | 4.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 2.6 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 1.4 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 3.0 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 1.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 4.7 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 2.1 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.1 | 1.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 2.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 2.4 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.1 | 2.4 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 4.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.4 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.1 | 0.9 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.1 | 1.3 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 | 1.6 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 | 2.6 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 10.8 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 1.8 | GO:0051084 | 'de novo' posttranslational protein folding(GO:0051084) |
| 0.1 | 12.1 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.1 | 0.9 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 0.3 | GO:0018874 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 0.1 | 1.3 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 3.0 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 | 2.8 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.1 | 0.5 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 0.8 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.1 | 7.0 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.1 | 0.1 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 1.0 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.1 | 1.2 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 4.0 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 3.2 | GO:0021795 | cerebral cortex cell migration(GO:0021795) |
| 0.1 | 2.7 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.1 | 1.0 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 6.4 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.1 | 2.6 | GO:0065002 | intracellular protein transmembrane transport(GO:0065002) |
| 0.1 | 1.6 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.1 | 1.6 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 0.5 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 2.1 | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) |
| 0.1 | 0.9 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 3.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 2.2 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 | 12.2 | GO:0006413 | translational initiation(GO:0006413) |
| 0.1 | 4.0 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.1 | 0.7 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 0.7 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 1.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.7 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 1.6 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 0.4 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 1.0 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.2 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.2 | GO:0051004 | regulation of lipoprotein lipase activity(GO:0051004) |
| 0.0 | 0.7 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.8 | GO:0043928 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 2.4 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.0 | 1.4 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.2 | GO:0070255 | regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.4 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 | 1.6 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.8 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 1.3 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.4 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.3 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 1.6 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 1.3 | GO:0030330 | DNA damage response, signal transduction by p53 class mediator(GO:0030330) |
| 0.0 | 0.5 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 1.0 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 1.9 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.0 | 0.1 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) negative regulation of muscle hypertrophy(GO:0014741) |
| 0.0 | 6.4 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.0 | 0.2 | GO:0051043 | regulation of membrane protein ectodomain proteolysis(GO:0051043) |
| 0.0 | 5.0 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 0.3 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.1 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 22.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 5.5 | 22.0 | GO:1990393 | 3M complex(GO:1990393) |
| 4.9 | 19.6 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 3.8 | 18.8 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 3.0 | 35.6 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 2.8 | 5.7 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 2.5 | 15.0 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 2.5 | 10.0 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 2.4 | 7.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 2.2 | 6.7 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 2.1 | 12.8 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 2.1 | 29.8 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 2.0 | 8.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 2.0 | 20.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 2.0 | 9.9 | GO:0097513 | myosin II filament(GO:0097513) |
| 1.9 | 9.4 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 1.7 | 10.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 1.5 | 9.2 | GO:0014802 | terminal cisterna(GO:0014802) |
| 1.5 | 25.0 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 1.3 | 5.4 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 1.3 | 7.9 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 1.3 | 20.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 1.2 | 18.5 | GO:0005861 | troponin complex(GO:0005861) |
| 1.1 | 10.3 | GO:0070938 | contractile ring(GO:0070938) |
| 1.1 | 11.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 1.1 | 11.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 1.1 | 5.5 | GO:0031673 | H zone(GO:0031673) |
| 1.1 | 3.3 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 1.1 | 21.3 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 1.0 | 2.9 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.9 | 42.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.9 | 17.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.9 | 4.6 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.9 | 5.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.9 | 13.7 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.8 | 7.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.7 | 17.1 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.7 | 2.9 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.7 | 34.5 | GO:0031430 | M band(GO:0031430) |
| 0.7 | 14.0 | GO:0043219 | lateral loop(GO:0043219) |
| 0.7 | 4.6 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.7 | 2.0 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.6 | 2.6 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.6 | 40.3 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.6 | 22.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.6 | 7.6 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.6 | 3.4 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.6 | 15.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.5 | 7.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.5 | 31.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.5 | 3.6 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic spine head(GO:0044327) |
| 0.5 | 8.1 | GO:0061202 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.5 | 5.0 | GO:0045180 | basal cortex(GO:0045180) |
| 0.5 | 4.0 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.5 | 8.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.5 | 8.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.5 | 9.5 | GO:0031672 | A band(GO:0031672) |
| 0.5 | 4.6 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.5 | 7.3 | GO:0045120 | pronucleus(GO:0045120) |
| 0.5 | 5.9 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.4 | 11.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.4 | 10.4 | GO:0043218 | compact myelin(GO:0043218) |
| 0.4 | 1.7 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.4 | 6.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.4 | 2.9 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.4 | 7.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.4 | 2.9 | GO:0043196 | varicosity(GO:0043196) |
| 0.4 | 7.5 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.4 | 6.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.4 | 1.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) outer acrosomal membrane(GO:0002081) |
| 0.4 | 16.4 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.4 | 5.4 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.3 | 0.3 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.3 | 2.9 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.3 | 3.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.3 | 19.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.3 | 1.5 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.3 | 14.5 | GO:0002102 | podosome(GO:0002102) |
| 0.3 | 16.6 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.3 | 3.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.3 | 0.8 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.3 | 2.5 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.3 | 4.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.3 | 0.3 | GO:0043260 | laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 0.3 | 1.3 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.3 | 9.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.3 | 2.7 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.3 | 4.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.3 | 14.5 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.3 | 6.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.3 | 4.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 7.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 18.9 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.2 | 36.2 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.2 | 7.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.2 | 6.6 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.2 | 17.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 2.8 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.2 | 2.8 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 7.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 0.7 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.2 | 3.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.2 | 12.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 1.4 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.2 | 2.8 | GO:0034709 | methylosome(GO:0034709) |
| 0.2 | 4.5 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.2 | 3.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 76.8 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.2 | 5.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 6.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 2.1 | GO:0033643 | host cell part(GO:0033643) |
| 0.2 | 5.8 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.2 | 13.5 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.2 | 3.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 0.7 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.2 | 1.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.2 | 2.4 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.2 | 53.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.2 | 0.5 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.2 | 0.3 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.2 | 28.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.2 | 0.8 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 4.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 2.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 3.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 2.4 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 1.0 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 2.5 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 9.3 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 2.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.5 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 14.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 1.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 1.7 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 8.7 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 10.6 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 1.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 3.7 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 1.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 2.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 0.4 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 4.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 17.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 40.1 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.1 | 1.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.2 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 2.4 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 1.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 3.4 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 0.9 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.1 | 4.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 28.2 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 3.4 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.1 | 2.6 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 1.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 1.6 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 8.0 | GO:0005938 | cell cortex(GO:0005938) |
| 0.1 | 1.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 5.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 0.4 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 2.5 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 2.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 0.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 1.0 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.4 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 1.5 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.1 | 1.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 4.9 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 0.3 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 2.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 21.5 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 1.1 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 3.8 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 4.8 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 1.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.7 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.2 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 14.5 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.7 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 3.3 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 2.0 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 1.7 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 0.2 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 1.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.9 | 38.8 | GO:0048030 | disaccharide binding(GO:0048030) |
| 6.3 | 25.0 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 4.9 | 19.6 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 4.5 | 13.4 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 4.3 | 12.9 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 4.2 | 12.6 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 3.8 | 11.4 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) bisphosphoglycerate phosphatase activity(GO:0034416) inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) |
| 3.0 | 15.0 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 2.7 | 8.2 | GO:0003974 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 2.7 | 10.9 | GO:0033265 | acetylcholinesterase activity(GO:0003990) choline binding(GO:0033265) |
| 2.7 | 8.1 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 2.5 | 10.0 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 2.5 | 9.8 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 2.3 | 9.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 2.1 | 6.2 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 2.1 | 12.5 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 2.1 | 6.2 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 2.0 | 6.0 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 1.9 | 5.8 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 1.9 | 13.4 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 1.8 | 9.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 1.8 | 14.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 1.8 | 41.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 1.7 | 10.5 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 1.7 | 13.9 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 1.7 | 23.6 | GO:0031014 | troponin T binding(GO:0031014) |
| 1.6 | 46.2 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 1.5 | 7.5 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 1.4 | 5.8 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 1.4 | 1.4 | GO:0031432 | titin binding(GO:0031432) RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 1.3 | 45.7 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 1.3 | 6.6 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 1.2 | 7.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 1.2 | 3.6 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 1.1 | 13.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 1.1 | 7.9 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 1.1 | 3.3 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 1.0 | 9.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 1.0 | 5.9 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 1.0 | 8.8 | GO:0045545 | syndecan binding(GO:0045545) |
| 1.0 | 18.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.9 | 6.6 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.9 | 7.4 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.9 | 13.8 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.9 | 8.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.9 | 3.6 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.9 | 12.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.9 | 2.6 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.8 | 20.0 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.8 | 17.1 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.8 | 3.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.8 | 2.4 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.8 | 11.0 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.8 | 8.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.8 | 14.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.8 | 7.8 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.8 | 14.0 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.8 | 5.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.8 | 3.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.8 | 5.3 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.7 | 2.9 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.7 | 2.9 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.7 | 8.6 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.7 | 25.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.7 | 4.8 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.7 | 3.4 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.7 | 3.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.7 | 58.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.7 | 11.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.7 | 5.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.6 | 2.6 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.6 | 19.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.6 | 3.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.6 | 3.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.6 | 2.4 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.6 | 1.8 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.6 | 4.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.6 | 12.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.6 | 2.4 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.6 | 6.4 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.6 | 3.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.6 | 4.5 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.6 | 5.0 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.6 | 12.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.5 | 1.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.5 | 1.6 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.5 | 10.3 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.5 | 3.8 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.5 | 3.7 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.5 | 4.8 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.5 | 2.6 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.5 | 4.7 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.5 | 3.6 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.5 | 11.0 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.5 | 25.5 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.5 | 2.3 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.5 | 1.8 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.5 | 11.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.5 | 3.6 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.4 | 2.2 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.4 | 17.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.4 | 1.3 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.4 | 21.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.4 | 11.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.4 | 22.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.4 | 1.6 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.4 | 5.3 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.4 | 6.9 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.4 | 4.8 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.4 | 16.7 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.4 | 6.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.4 | 10.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.4 | 4.2 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.4 | 12.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.4 | 2.6 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.4 | 1.9 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.4 | 2.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.4 | 4.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.4 | 2.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.4 | 3.3 | GO:0089720 | caspase binding(GO:0089720) |
| 0.4 | 4.0 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.4 | 8.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.4 | 2.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.4 | 7.8 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.3 | 16.0 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.3 | 4.2 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.3 | 1.3 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.3 | 5.3 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.3 | 1.6 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.3 | 1.9 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.3 | 2.9 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.3 | 1.6 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.3 | 2.5 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.3 | 3.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.3 | 12.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.3 | 4.9 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.3 | 1.5 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.3 | 13.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.3 | 6.5 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.3 | 1.8 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.3 | 19.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.3 | 13.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.3 | 1.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.3 | 4.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.3 | 44.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.3 | 4.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.3 | 2.4 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.3 | 1.3 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.3 | 2.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.3 | 2.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.3 | 12.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.3 | 0.8 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.3 | 1.0 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.2 | 5.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 4.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 0.7 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.2 | 0.7 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.2 | 3.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 4.7 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.2 | 22.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 10.3 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.2 | 3.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.2 | 8.9 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.2 | 0.9 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.2 | 3.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 3.9 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.2 | 3.9 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.2 | 2.6 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.2 | 5.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.2 | 2.5 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 3.6 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.2 | 1.0 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.2 | 0.6 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.2 | 6.0 | GO:0005416 | sodium:amino acid symporter activity(GO:0005283) cation:amino acid symporter activity(GO:0005416) |
| 0.2 | 1.8 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.2 | 3.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 2.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.2 | 1.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.2 | 0.9 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.2 | 1.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 4.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 5.9 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.2 | 6.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 0.5 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.2 | 2.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.2 | 19.0 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.2 | 1.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.2 | 6.9 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.2 | 2.8 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.2 | 29.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 3.0 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 2.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.6 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.7 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 1.7 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 2.3 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 1.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 1.2 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 2.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 4.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 1.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.9 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) phosphatidylinositol trisphosphate phosphatase activity(GO:0034594) |
| 0.1 | 0.8 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 0.6 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 0.9 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 1.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 1.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 14.6 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 0.7 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 3.1 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 12.2 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 2.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 0.6 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 3.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 1.5 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 1.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 0.4 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.1 | 5.5 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.1 | 1.4 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.1 | 27.0 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 40.9 | GO:0005525 | GTP binding(GO:0005525) |
| 0.1 | 2.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.5 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 4.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 4.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 1.1 | GO:0015662 | ATPase activity, coupled to transmembrane movement of ions, phosphorylative mechanism(GO:0015662) |
| 0.1 | 6.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.7 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 1.4 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.1 | 2.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 3.7 | GO:0004950 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.1 | 0.7 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 2.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 2.9 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 24.6 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.1 | 4.7 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.1 | 0.3 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.8 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 2.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.5 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.1 | 0.4 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.1 | 2.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.4 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 1.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.6 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 1.2 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 7.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 7.4 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 12.6 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.7 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.4 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.3 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 0.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.4 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 1.4 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.4 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 1.8 | GO:0008083 | growth factor activity(GO:0008083) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 26.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.9 | 18.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.8 | 10.3 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.6 | 36.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.6 | 8.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.5 | 19.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.5 | 3.0 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.4 | 25.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.4 | 32.7 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.4 | 29.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.4 | 12.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.4 | 17.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.3 | 18.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.3 | 35.5 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.3 | 10.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.3 | 13.8 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.3 | 33.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.3 | 12.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.3 | 27.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.3 | 18.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.3 | 5.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.3 | 14.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.2 | 4.0 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 13.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 5.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 7.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.2 | 5.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.2 | 2.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 5.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.2 | 12.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 31.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 3.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.2 | 10.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.2 | 4.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 6.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 10.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 2.7 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 6.0 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 0.8 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 1.7 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 0.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 2.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 6.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 4.6 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 1.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 1.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.3 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.3 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.4 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.2 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 67.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 1.0 | 35.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.8 | 13.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.8 | 17.8 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.8 | 29.0 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.8 | 7.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.8 | 16.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.7 | 21.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.7 | 11.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.7 | 15.8 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.6 | 7.7 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.6 | 15.7 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.6 | 12.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.6 | 21.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.6 | 12.0 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.6 | 19.3 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.5 | 12.5 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.5 | 2.7 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.5 | 8.5 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.5 | 18.2 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.4 | 15.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.4 | 10.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.4 | 8.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.4 | 4.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.3 | 13.1 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.3 | 8.1 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.3 | 7.5 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 5.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.3 | 9.1 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.3 | 8.8 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.3 | 7.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.3 | 22.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.3 | 5.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 10.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 4.1 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.3 | 12.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.3 | 26.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.3 | 3.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 9.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 3.9 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.2 | 9.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 5.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 12.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 2.6 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.2 | 13.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 8.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 0.6 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.2 | 3.7 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 14.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 11.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.2 | 7.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.2 | 6.2 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.2 | 5.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.2 | 2.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.2 | 6.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.2 | 2.1 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.2 | 1.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.2 | 10.7 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 1.5 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.1 | 10.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 0.7 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 3.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 2.9 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 4.7 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 1.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 2.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 3.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 3.8 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 1.9 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 9.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 0.4 | REACTOME G PROTEIN BETA GAMMA SIGNALLING | Genes involved in G-protein beta:gamma signalling |
| 0.1 | 0.7 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 5.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 11.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 2.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 1.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 0.8 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 6.0 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.1 | 1.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 1.3 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 3.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 4.5 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.8 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.6 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.6 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 1.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.6 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 3.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 2.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.4 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 2.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |