Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
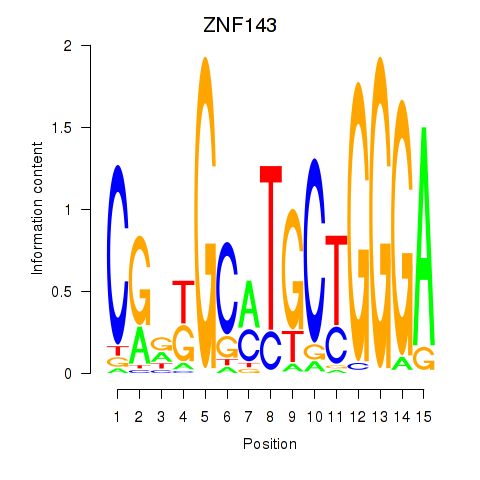
Results for ZNF143
Z-value: 3.83
Transcription factors associated with ZNF143
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF143
|
ENSG00000166478.10 | ZNF143 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF143 | hg38_v1_chr11_+_9461003_9461057 | 0.65 | 1.6e-27 | Click! |
Activity profile of ZNF143 motif
Sorted Z-values of ZNF143 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF143
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 51.2 | 153.5 | GO:0075732 | viral penetration into host nucleus(GO:0075732) multi-organism nuclear import(GO:1902594) |
| 41.1 | 123.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 35.7 | 250.2 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 33.5 | 100.6 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) mitochondrial membrane fission(GO:0090149) |
| 25.7 | 77.0 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 22.6 | 113.0 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 21.8 | 65.4 | GO:1903778 | positive regulation of histamine secretion by mast cell(GO:1903595) protein localization to vacuolar membrane(GO:1903778) |
| 21.5 | 150.6 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 20.9 | 83.7 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 20.1 | 120.6 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 20.1 | 120.4 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 19.2 | 134.7 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 18.3 | 54.9 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 18.1 | 54.4 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 17.2 | 120.5 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 17.0 | 102.0 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 16.8 | 84.0 | GO:0039019 | posttranslational protein targeting to membrane(GO:0006620) pronephric nephron development(GO:0039019) |
| 15.0 | 119.7 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 14.8 | 207.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 14.7 | 162.0 | GO:0036481 | intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036481) |
| 14.6 | 131.5 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 14.3 | 57.1 | GO:0006272 | leading strand elongation(GO:0006272) |
| 13.5 | 67.7 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 13.2 | 105.9 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 12.7 | 51.0 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 12.2 | 195.7 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 11.7 | 46.7 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 11.7 | 35.0 | GO:0097252 | negative regulation of helicase activity(GO:0051097) oligodendrocyte apoptotic process(GO:0097252) |
| 11.6 | 115.8 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 11.6 | 34.7 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 11.2 | 134.9 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 11.0 | 33.0 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 10.8 | 54.2 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 10.6 | 127.8 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 10.6 | 63.3 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 10.1 | 50.4 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 9.5 | 114.3 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 9.3 | 56.0 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 9.2 | 46.1 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 8.8 | 35.2 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 8.7 | 52.4 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 8.5 | 33.8 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 8.3 | 116.3 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 8.2 | 32.9 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 8.1 | 32.5 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 7.7 | 100.7 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 7.4 | 44.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 7.3 | 73.3 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 7.2 | 57.8 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 7.2 | 65.0 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 7.1 | 21.3 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 7.0 | 48.7 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 6.9 | 20.7 | GO:0070512 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 6.9 | 55.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 6.9 | 13.7 | GO:0048254 | snoRNA localization(GO:0048254) |
| 6.8 | 27.3 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 6.8 | 81.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 6.7 | 26.9 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 6.5 | 39.3 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 6.5 | 58.3 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 6.3 | 18.8 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 6.2 | 86.3 | GO:1904871 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 6.1 | 24.3 | GO:1901377 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 6.0 | 42.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 6.0 | 18.0 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 6.0 | 29.8 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 5.9 | 23.7 | GO:0046778 | modification by virus of host mRNA processing(GO:0046778) |
| 5.9 | 29.4 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 5.8 | 17.5 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 5.8 | 17.5 | GO:1900110 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) |
| 5.7 | 34.3 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 5.7 | 39.8 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 5.7 | 17.0 | GO:0000967 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 5.6 | 45.0 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 5.5 | 44.0 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 5.3 | 64.0 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 5.3 | 26.5 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 5.0 | 100.6 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 5.0 | 65.2 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 5.0 | 75.1 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 4.9 | 49.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 4.8 | 24.2 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 4.8 | 67.5 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 4.5 | 22.4 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) |
| 4.4 | 182.3 | GO:0071174 | mitotic spindle checkpoint(GO:0071174) |
| 4.4 | 13.1 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 4.4 | 87.1 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 4.1 | 78.8 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 4.1 | 16.3 | GO:1904116 | traversing start control point of mitotic cell cycle(GO:0007089) response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 4.0 | 64.6 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 3.9 | 89.7 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 3.8 | 11.5 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 3.8 | 98.8 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 3.7 | 11.1 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 3.6 | 29.1 | GO:0030091 | protein repair(GO:0030091) |
| 3.6 | 21.3 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 3.5 | 17.7 | GO:2000814 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 3.5 | 34.5 | GO:0016074 | snoRNA metabolic process(GO:0016074) snoRNA processing(GO:0043144) |
| 3.4 | 30.9 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 3.4 | 54.7 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 3.3 | 16.7 | GO:0030242 | pexophagy(GO:0030242) |
| 3.3 | 13.2 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 3.2 | 22.5 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 3.1 | 21.9 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 3.1 | 9.3 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 3.1 | 18.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 3.1 | 6.2 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 3.1 | 12.3 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 3.1 | 45.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 3.0 | 39.6 | GO:0016180 | snRNA processing(GO:0016180) |
| 3.0 | 12.0 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 2.9 | 23.5 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 2.9 | 26.3 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 2.9 | 34.9 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 2.9 | 191.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 2.9 | 34.8 | GO:0045116 | protein neddylation(GO:0045116) |
| 2.9 | 5.8 | GO:1903405 | regulation of establishment of protein localization to telomere(GO:0070203) protein localization to nuclear body(GO:1903405) positive regulation of establishment of protein localization to telomere(GO:1904851) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 2.7 | 32.9 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 2.7 | 13.7 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 2.7 | 92.1 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 2.7 | 56.9 | GO:0031167 | rRNA methylation(GO:0031167) |
| 2.7 | 13.4 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 2.7 | 16.0 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 2.6 | 18.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 2.6 | 15.5 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) positive regulation of t-circle formation(GO:1904431) |
| 2.4 | 46.5 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 2.4 | 33.9 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 2.4 | 60.2 | GO:0051031 | tRNA export from nucleus(GO:0006409) tRNA transport(GO:0051031) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 2.4 | 16.7 | GO:0009249 | protein lipoylation(GO:0009249) |
| 2.4 | 71.4 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 2.4 | 37.9 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 2.4 | 23.6 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 2.4 | 11.8 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 2.3 | 9.2 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 2.3 | 20.6 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 2.3 | 4.6 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 2.3 | 13.5 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 2.2 | 83.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 2.2 | 17.2 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 2.2 | 15.1 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 2.1 | 17.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 2.1 | 58.0 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 2.1 | 58.8 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 2.0 | 26.3 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 2.0 | 10.0 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 2.0 | 62.0 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 2.0 | 55.7 | GO:0006465 | signal peptide processing(GO:0006465) |
| 2.0 | 11.7 | GO:0044211 | CTP salvage(GO:0044211) |
| 1.9 | 57.6 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 1.9 | 42.2 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 1.9 | 13.3 | GO:0031133 | cellular magnesium ion homeostasis(GO:0010961) regulation of axon diameter(GO:0031133) |
| 1.9 | 36.0 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 1.9 | 19.0 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 1.9 | 18.9 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 1.8 | 72.1 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 1.8 | 9.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 1.8 | 23.7 | GO:0009112 | nucleobase metabolic process(GO:0009112) |
| 1.8 | 23.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 1.8 | 16.3 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 1.8 | 349.3 | GO:0006364 | rRNA processing(GO:0006364) |
| 1.8 | 33.4 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 1.6 | 14.8 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 1.6 | 8.2 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 1.5 | 27.5 | GO:0009452 | 7-methylguanosine mRNA capping(GO:0006370) 7-methylguanosine RNA capping(GO:0009452) RNA capping(GO:0036260) |
| 1.5 | 33.4 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 1.5 | 25.5 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 1.5 | 28.5 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 1.5 | 16.2 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 1.4 | 32.9 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 1.4 | 475.1 | GO:0000375 | RNA splicing, via transesterification reactions(GO:0000375) |
| 1.4 | 12.4 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 1.3 | 18.9 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 1.3 | 4.0 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 1.3 | 54.2 | GO:0035872 | nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway(GO:0035872) nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) |
| 1.3 | 42.6 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 1.3 | 14.1 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 1.2 | 6.1 | GO:0003069 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 1.2 | 24.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 1.2 | 14.6 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 1.2 | 8.5 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 1.2 | 135.6 | GO:0008380 | RNA splicing(GO:0008380) |
| 1.2 | 15.4 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 1.2 | 5.9 | GO:0071609 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) chemokine (C-C motif) ligand 5 production(GO:0071609) |
| 1.2 | 14.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 1.2 | 10.5 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 1.2 | 6.9 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 1.2 | 9.2 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 1.1 | 103.3 | GO:1904667 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 1.1 | 28.2 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 1.1 | 15.4 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 1.0 | 27.0 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 1.0 | 65.5 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.9 | 18.9 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.9 | 3.6 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.9 | 10.7 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.9 | 24.8 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.9 | 10.5 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.9 | 6.1 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.9 | 12.8 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.8 | 6.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.8 | 29.6 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.8 | 3.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.8 | 13.5 | GO:0048535 | lymph node development(GO:0048535) |
| 0.7 | 139.9 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.7 | 4.1 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.7 | 5.4 | GO:0090656 | t-circle formation(GO:0090656) |
| 0.6 | 48.7 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.6 | 14.1 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.6 | 42.4 | GO:0006413 | translational initiation(GO:0006413) |
| 0.6 | 5.0 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.6 | 44.2 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.5 | 2.1 | GO:0051299 | mitotic centrosome separation(GO:0007100) centrosome separation(GO:0051299) |
| 0.5 | 13.2 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.5 | 3.7 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.5 | 3.6 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.4 | 11.2 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.4 | 22.6 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.4 | 37.8 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.4 | 17.2 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.4 | 20.6 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.4 | 29.1 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.4 | 4.1 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.4 | 11.6 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.4 | 1.2 | GO:1904579 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) response to hypobaric hypoxia(GO:1990910) |
| 0.4 | 11.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.4 | 5.7 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.4 | 8.5 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.3 | 1.7 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.3 | 26.5 | GO:0048678 | response to axon injury(GO:0048678) |
| 0.3 | 5.7 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.3 | 6.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.3 | 33.2 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.3 | 15.7 | GO:0017145 | stem cell division(GO:0017145) |
| 0.3 | 1.8 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.3 | 3.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) negative regulation of sprouting angiogenesis(GO:1903671) |
| 0.3 | 15.2 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.3 | 3.8 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.3 | 18.8 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.2 | 18.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.2 | 2.6 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.2 | 15.4 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.2 | 28.6 | GO:0046546 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.2 | 23.8 | GO:0007613 | memory(GO:0007613) |
| 0.2 | 3.6 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.2 | 2.4 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.2 | 5.6 | GO:0036344 | platelet morphogenesis(GO:0036344) |
| 0.2 | 0.6 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 2.5 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 0.9 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 4.3 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.1 | 27.4 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.1 | 3.1 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 6.6 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 7.3 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.1 | 7.0 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 2.7 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.1 | 6.3 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.1 | 1.0 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 8.6 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.0 | 2.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 6.5 | GO:0042113 | B cell activation(GO:0042113) |
| 0.0 | 2.1 | GO:0030879 | mammary gland development(GO:0030879) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 31.9 | 95.8 | GO:0033597 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 30.2 | 271.6 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 25.7 | 77.0 | GO:0018444 | translation release factor complex(GO:0018444) |
| 22.9 | 68.7 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 19.9 | 199.3 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 19.2 | 96.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 18.6 | 93.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 16.8 | 168.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 16.7 | 83.7 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 15.0 | 45.0 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 14.8 | 103.5 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 14.2 | 42.7 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 13.9 | 83.6 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 13.5 | 134.9 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 13.1 | 78.8 | GO:0005816 | spindle pole body(GO:0005816) |
| 11.6 | 34.9 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 11.4 | 57.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 11.3 | 33.9 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 11.1 | 44.3 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 10.9 | 65.4 | GO:0070545 | PeBoW complex(GO:0070545) |
| 10.6 | 170.1 | GO:0034709 | methylosome(GO:0034709) |
| 10.5 | 147.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 10.2 | 71.2 | GO:0031415 | NatA complex(GO:0031415) |
| 9.9 | 49.5 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 9.7 | 87.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 9.7 | 67.7 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 9.5 | 28.5 | GO:0043260 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 9.3 | 93.5 | GO:0070552 | BRISC complex(GO:0070552) |
| 8.8 | 88.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 8.8 | 26.3 | GO:0034455 | t-UTP complex(GO:0034455) |
| 8.6 | 94.7 | GO:0070449 | elongin complex(GO:0070449) |
| 8.4 | 42.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 8.4 | 67.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 8.3 | 33.4 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 8.3 | 141.8 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 7.3 | 58.3 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 7.1 | 64.0 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 7.1 | 77.9 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 7.1 | 91.9 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 7.0 | 56.0 | GO:0000796 | condensin complex(GO:0000796) |
| 6.9 | 20.8 | GO:0030689 | Noc complex(GO:0030689) |
| 6.9 | 20.6 | GO:0034657 | GID complex(GO:0034657) |
| 6.9 | 27.5 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 6.6 | 86.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 6.5 | 32.5 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 6.2 | 24.8 | GO:0071986 | Ragulator complex(GO:0071986) |
| 6.2 | 55.7 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 6.2 | 61.8 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 5.6 | 67.5 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 5.4 | 27.0 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 5.4 | 16.1 | GO:1903349 | omegasome membrane(GO:1903349) |
| 5.3 | 398.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 5.2 | 62.8 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 5.1 | 91.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 5.1 | 131.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 4.9 | 14.8 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 4.9 | 58.8 | GO:0090543 | F-actin capping protein complex(GO:0008290) Flemming body(GO:0090543) |
| 4.7 | 33.0 | GO:0090661 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 4.3 | 85.8 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 4.1 | 20.7 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 4.1 | 65.4 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 4.0 | 24.2 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 4.0 | 19.9 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 3.9 | 58.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 3.8 | 18.9 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 3.7 | 44.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 3.7 | 99.8 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 3.6 | 18.2 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 3.6 | 39.6 | GO:0032039 | integrator complex(GO:0032039) |
| 3.6 | 39.3 | GO:0005638 | lamin filament(GO:0005638) |
| 3.4 | 13.7 | GO:0005687 | U4 snRNP(GO:0005687) |
| 3.4 | 6.8 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 3.3 | 40.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 3.0 | 82.9 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 2.9 | 46.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 2.9 | 11.7 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 2.9 | 17.5 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 2.9 | 84.6 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 2.8 | 51.0 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 2.8 | 277.3 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 2.7 | 57.4 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 2.7 | 34.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 2.7 | 8.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 2.4 | 220.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 2.3 | 9.3 | GO:1990423 | RZZ complex(GO:1990423) |
| 2.3 | 133.8 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 2.3 | 43.0 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 2.2 | 60.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 2.2 | 15.4 | GO:1990130 | Iml1 complex(GO:1990130) |
| 2.2 | 17.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 2.2 | 13.0 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 2.1 | 41.2 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 2.0 | 8.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 2.0 | 4.0 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 2.0 | 31.9 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 1.9 | 15.5 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) ERCC4-ERCC1 complex(GO:0070522) |
| 1.9 | 108.2 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 1.8 | 57.2 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 1.8 | 153.1 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 1.7 | 11.6 | GO:0016272 | prefoldin complex(GO:0016272) |
| 1.6 | 22.2 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 1.5 | 150.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 1.4 | 80.8 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 1.4 | 75.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 1.4 | 15.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 1.4 | 81.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 1.4 | 108.1 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 1.3 | 42.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 1.3 | 100.6 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 1.3 | 81.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 1.3 | 113.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 1.2 | 74.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 1.0 | 41.4 | GO:0016592 | mediator complex(GO:0016592) |
| 1.0 | 9.9 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.9 | 18.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.9 | 18.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.9 | 39.4 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.9 | 103.4 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.8 | 21.1 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.8 | 31.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.8 | 7.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.7 | 19.0 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.7 | 87.0 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.7 | 15.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.7 | 92.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.6 | 53.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.6 | 23.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.6 | 15.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.6 | 37.2 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.6 | 5.0 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.5 | 7.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.5 | 8.7 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.5 | 18.8 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.4 | 67.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.4 | 354.8 | GO:0005730 | nucleolus(GO:0005730) |
| 0.4 | 2.8 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.4 | 17.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.4 | 7.6 | GO:0030904 | retromer complex(GO:0030904) |
| 0.4 | 10.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.4 | 7.3 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.3 | 4.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.3 | 26.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.3 | 93.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.3 | 4.9 | GO:0071437 | invadopodium(GO:0071437) |
| 0.3 | 54.8 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.3 | 22.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.3 | 5.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.3 | 10.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 4.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 3.1 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 2.5 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 3.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 1.7 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 5.8 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 1.2 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 3.6 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 3.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 41.1 | 123.3 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 40.2 | 120.5 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 22.4 | 134.7 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 21.3 | 127.8 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 20.9 | 62.8 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 20.6 | 82.6 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 19.9 | 199.3 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 19.2 | 96.2 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 19.0 | 57.1 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 19.0 | 56.9 | GO:0008988 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) |
| 18.3 | 54.9 | GO:0003896 | DNA primase activity(GO:0003896) |
| 17.0 | 102.0 | GO:0004793 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 16.8 | 50.5 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 16.7 | 83.7 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 15.5 | 124.0 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 14.7 | 44.0 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 14.1 | 42.2 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 13.1 | 39.3 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 12.7 | 50.9 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 11.2 | 134.9 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 11.0 | 77.0 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 10.0 | 90.0 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 9.9 | 59.7 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 9.9 | 79.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 9.6 | 57.6 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 9.5 | 56.9 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) 5'-flap endonuclease activity(GO:0017108) |
| 9.3 | 37.2 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 9.0 | 81.3 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 8.5 | 93.6 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 8.4 | 50.4 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 8.2 | 32.9 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 8.1 | 65.0 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 8.0 | 48.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 8.0 | 63.7 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 7.8 | 46.7 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 7.7 | 99.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 7.2 | 143.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 7.1 | 213.9 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 6.9 | 193.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 6.8 | 20.5 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 6.8 | 54.7 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 6.3 | 18.9 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 6.2 | 74.7 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 6.2 | 87.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 6.0 | 36.0 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 5.6 | 28.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 5.3 | 31.9 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 5.3 | 100.6 | GO:0051400 | BH domain binding(GO:0051400) |
| 5.3 | 47.6 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 5.3 | 283.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 5.1 | 71.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 5.0 | 35.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 4.7 | 33.0 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 4.6 | 100.7 | GO:0008494 | translation activator activity(GO:0008494) |
| 4.3 | 17.2 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 4.3 | 21.3 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 4.2 | 150.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 3.9 | 35.0 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 3.8 | 45.8 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 3.8 | 11.4 | GO:0009032 | thymidine phosphorylase activity(GO:0009032) pyrimidine-nucleoside phosphorylase activity(GO:0016154) |
| 3.7 | 14.8 | GO:0002046 | opsin binding(GO:0002046) |
| 3.5 | 31.8 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 3.3 | 39.9 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 3.3 | 16.6 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 3.1 | 62.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 3.0 | 23.6 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 2.9 | 111.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 2.9 | 29.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 2.8 | 47.0 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 2.7 | 32.9 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 2.7 | 133.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 2.7 | 10.7 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 2.7 | 8.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 2.7 | 23.9 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 2.6 | 57.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 2.6 | 96.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 2.6 | 12.8 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 2.6 | 173.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 2.5 | 17.5 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 2.3 | 27.3 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 2.3 | 6.8 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 2.2 | 32.5 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 2.1 | 6.2 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 2.1 | 4.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 2.0 | 82.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 2.0 | 16.0 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 2.0 | 11.7 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 2.0 | 13.7 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 2.0 | 35.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 1.9 | 15.5 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 1.9 | 15.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 1.8 | 51.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 1.7 | 18.6 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 1.7 | 5.0 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 1.6 | 11.5 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 1.6 | 47.2 | GO:0071949 | FAD binding(GO:0071949) |
| 1.5 | 47.8 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 1.5 | 12.3 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 1.4 | 95.2 | GO:0008235 | aminopeptidase activity(GO:0004177) metalloexopeptidase activity(GO:0008235) |
| 1.4 | 63.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 1.4 | 15.1 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 1.3 | 5.2 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 1.3 | 13.0 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 1.3 | 22.7 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 1.3 | 42.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 1.2 | 264.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 1.2 | 60.6 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 1.2 | 6.0 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 1.2 | 18.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 1.2 | 10.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 1.1 | 23.8 | GO:0030515 | snoRNA binding(GO:0030515) |
| 1.1 | 8.6 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 1.1 | 48.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 1.0 | 4.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 1.0 | 141.4 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 1.0 | 169.1 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 1.0 | 84.0 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 1.0 | 34.9 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.9 | 18.9 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.9 | 20.6 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.9 | 5.5 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.9 | 51.2 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.9 | 23.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.8 | 22.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.8 | 11.8 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.7 | 75.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.7 | 16.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.7 | 26.5 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.7 | 24.3 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.7 | 5.4 | GO:0036310 | annealing helicase activity(GO:0036310) annealing activity(GO:0097617) |
| 0.7 | 7.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.7 | 17.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.6 | 68.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.6 | 8.9 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.6 | 13.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.6 | 6.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.6 | 3.6 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.6 | 16.2 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.6 | 32.0 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.6 | 6.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.5 | 11.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.5 | 160.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.5 | 759.4 | GO:0003723 | RNA binding(GO:0003723) |
| 0.5 | 8.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.5 | 2.6 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.5 | 39.6 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.5 | 4.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.4 | 41.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.4 | 4.7 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.3 | 3.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.3 | 19.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.3 | 12.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.3 | 5.6 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.3 | 3.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.3 | 20.9 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.3 | 24.5 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.3 | 31.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.3 | 4.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.3 | 25.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.3 | 44.7 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.3 | 15.5 | GO:0043621 | protein self-association(GO:0043621) |
| 0.2 | 38.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.2 | 3.7 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.2 | 11.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 10.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.2 | 2.6 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.2 | 1.8 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.2 | 7.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 31.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.2 | 16.6 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.2 | 82.8 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.2 | 3.7 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 9.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.9 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 4.0 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.1 | 3.3 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 1.7 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.1 | 4.3 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.1 | 2.4 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 1.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 1.4 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 2.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 2.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.8 | GO:0015485 | cholesterol binding(GO:0015485) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 363.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 3.6 | 103.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 3.3 | 13.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 3.3 | 130.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 2.9 | 144.0 | PID MYC PATHWAY | C-MYC pathway |
| 2.3 | 37.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 2.3 | 52.8 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 1.8 | 138.3 | PID P53 REGULATION PATHWAY | p53 pathway |
| 1.5 | 28.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 1.4 | 131.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 1.2 | 62.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 1.2 | 137.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 1.1 | 70.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 1.1 | 42.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 1.1 | 81.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.9 | 20.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.9 | 29.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.8 | 81.2 | PID P73PATHWAY | p73 transcription factor network |
| 0.8 | 29.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.7 | 22.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.7 | 111.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.7 | 45.0 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.5 | 6.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.4 | 14.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.3 | 19.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.3 | 19.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.3 | 6.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 17.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 10.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.2 | 3.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.2 | 5.6 | PID FOXO PATHWAY | FoxO family signaling |
| 0.2 | 9.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.2 | 8.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 3.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 5.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 4.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 3.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 4.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.4 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 4.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.7 | 35.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 11.1 | 232.6 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 8.9 | 44.3 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 8.2 | 16.3 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 6.7 | 127.8 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 6.3 | 232.2 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 6.1 | 171.3 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 5.7 | 34.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 5.6 | 163.3 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 5.3 | 84.0 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 4.5 | 439.4 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 4.1 | 115.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 4.1 | 82.3 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 4.1 | 106.7 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 4.1 | 65.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 4.0 | 96.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 4.0 | 179.3 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 3.6 | 57.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 3.6 | 118.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 3.2 | 54.2 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 3.1 | 74.7 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 2.9 | 55.7 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 2.9 | 204.7 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 2.7 | 86.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 2.7 | 229.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 2.4 | 40.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 2.3 | 36.0 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 2.2 | 22.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 2.1 | 45.0 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 2.1 | 10.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 1.9 | 28.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 1.8 | 54.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 1.8 | 42.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 1.7 | 43.0 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 1.7 | 77.1 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 1.7 | 198.5 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 1.6 | 50.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 1.6 | 108.4 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 1.6 | 155.2 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 1.6 | 18.8 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 1.6 | 78.1 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 1.6 | 110.4 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 1.5 | 136.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 1.5 | 46.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 1.5 | 17.7 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 1.5 | 118.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 1.4 | 31.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 1.4 | 7.0 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 1.3 | 33.6 | REACTOME TRANSLATION | Genes involved in Translation |
| 1.2 | 11.6 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 1.1 | 17.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 1.1 | 9.6 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 1.0 | 17.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 1.0 | 15.1 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 1.0 | 16.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 1.0 | 26.0 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.9 | 50.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.9 | 22.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.8 | 33.9 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.8 | 11.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.7 | 130.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.7 | 28.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.6 | 16.8 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.6 | 18.9 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.5 | 23.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.5 | 71.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.5 | 8.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.4 | 6.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.3 | 5.6 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.3 | 5.8 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.3 | 28.5 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.2 | 10.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.2 | 2.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 14.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 7.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 11.5 | REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |
| 0.1 | 6.0 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 2.8 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 1.4 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 3.4 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 1.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |