Project
GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
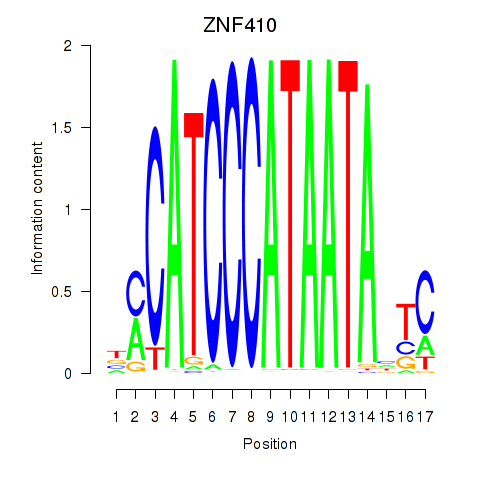
Results for ZNF410
Z-value: 0.37
Transcription factors associated with ZNF410
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF410
|
ENSG00000119725.20 | ZNF410 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF410 | hg38_v1_chr14_+_73886617_73886766 | 0.30 | 4.7e-06 | Click! |
Activity profile of ZNF410 motif
Sorted Z-values of ZNF410 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF410
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_6607334 | 23.03 |
ENST00000645645.1
ENST00000357008.7 ENST00000544484.6 ENST00000544040.7 |
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr12_-_6607397 | 21.21 |
ENST00000645005.1
ENST00000646806.1 |
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr21_+_43565176 | 4.95 |
ENST00000599962.2
|
H2BS1
|
H2B.S histone 1 |
| chr14_-_21383989 | 4.44 |
ENST00000216297.7
|
SUPT16H
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr21_-_5973383 | 4.40 |
ENST00000464664.3
|
ENSG00000274559.3
|
novel histone H2B family protein |
| chr11_-_57567617 | 3.67 |
ENST00000287156.9
|
UBE2L6
|
ubiquitin conjugating enzyme E2 L6 |
| chr12_+_121803991 | 3.19 |
ENST00000604567.6
ENST00000542440.5 |
SETD1B
|
SET domain containing 1B, histone lysine methyltransferase |
| chr22_-_38872206 | 1.98 |
ENST00000407418.8
ENST00000216083.6 |
CBX6
|
chromobox 6 |
| chr2_+_69893940 | 1.40 |
ENST00000244227.8
ENST00000409116.5 |
SNRNP27
|
small nuclear ribonucleoprotein U4/U6.U5 subunit 27 |
| chr1_+_206203541 | 0.17 |
ENST00000573034.8
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 44.2 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.5 | 3.7 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.2 | 4.4 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 3.2 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.2 | GO:0051014 | actin filament severing(GO:0051014) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 44.2 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 3.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 4.4 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 1.5 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 44.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.7 | 3.7 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.2 | 4.4 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.2 | 3.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 1.5 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 44.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.4 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 3.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |