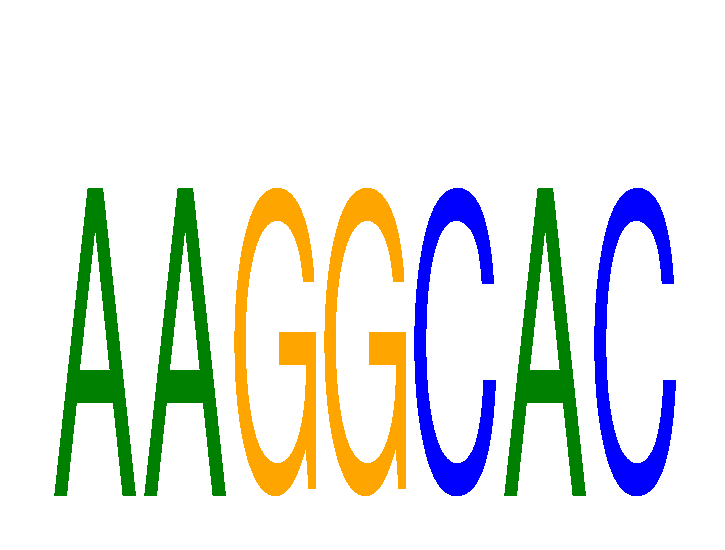Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for AAGGCAC
Z-value: 1.03
miRNA associated with seed AAGGCAC
| Name | miRBASE accession |
|---|---|
|
hsa-miR-124-3p.1
|
MIMAT0000422 |
Activity profile of AAGGCAC motif
Sorted Z-values of AAGGCAC motif
Network of associatons between targets according to the STRING database.
First level regulatory network of AAGGCAC
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.8 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 4.1 | 12.2 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 3.3 | 9.8 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 3.2 | 9.5 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 3.0 | 15.1 | GO:0032792 | negative regulation of triglyceride biosynthetic process(GO:0010868) negative regulation of CREB transcription factor activity(GO:0032792) |
| 2.9 | 8.7 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 2.6 | 10.2 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 2.5 | 10.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 2.4 | 7.3 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 2.4 | 16.8 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) regulation of collagen catabolic process(GO:0010710) |
| 2.4 | 19.1 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 2.4 | 9.6 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 2.3 | 7.0 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 2.3 | 9.2 | GO:0035740 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 2.3 | 6.9 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 2.3 | 9.0 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 2.2 | 8.8 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 2.2 | 6.6 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 2.1 | 6.3 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 2.0 | 6.1 | GO:0060435 | bronchiole development(GO:0060435) |
| 2.0 | 6.1 | GO:0032912 | negative regulation of transforming growth factor beta2 production(GO:0032912) |
| 2.0 | 6.1 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 2.0 | 11.8 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 1.9 | 5.8 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 1.8 | 5.5 | GO:0036510 | trimming of terminal mannose on C branch(GO:0036510) |
| 1.8 | 7.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 1.7 | 5.2 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 1.7 | 8.6 | GO:1900756 | protein processing in phagocytic vesicle(GO:1900756) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 1.7 | 23.6 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 1.7 | 6.6 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 1.6 | 1.6 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 1.6 | 8.1 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 1.5 | 6.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 1.5 | 4.5 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 1.5 | 11.8 | GO:0032803 | regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) |
| 1.5 | 10.2 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 1.5 | 14.6 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 1.4 | 10.0 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 1.4 | 5.7 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) negative regulation of glial cell migration(GO:1903976) |
| 1.3 | 4.0 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 1.3 | 3.9 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 1.3 | 5.2 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 1.3 | 3.8 | GO:1903249 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 1.2 | 5.0 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 1.2 | 8.7 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 1.2 | 4.6 | GO:0061552 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) endothelial tip cell fate specification(GO:0097102) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 1.1 | 5.7 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 1.1 | 4.6 | GO:1904502 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 1.1 | 2.3 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 1.1 | 6.8 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 1.1 | 5.6 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 1.1 | 6.7 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 1.1 | 5.5 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 1.1 | 3.3 | GO:1903772 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) regulation of viral budding via host ESCRT complex(GO:1903772) |
| 1.1 | 3.3 | GO:1902724 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 1.1 | 8.6 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 1.1 | 8.5 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 1.1 | 1.1 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 1.0 | 4.1 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) |
| 1.0 | 6.1 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) mRNA cleavage involved in gene silencing(GO:0098795) |
| 1.0 | 4.0 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 1.0 | 2.0 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 1.0 | 3.9 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 1.0 | 2.9 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 1.0 | 13.5 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.9 | 5.7 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.9 | 2.8 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.9 | 8.4 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.9 | 1.8 | GO:0036166 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.9 | 2.7 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.9 | 3.5 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.9 | 24.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.9 | 2.6 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.8 | 15.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.8 | 6.6 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.8 | 2.4 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.8 | 6.4 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.8 | 4.8 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.8 | 2.4 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.8 | 3.2 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.8 | 2.3 | GO:0060367 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 0.8 | 3.1 | GO:0018032 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 0.8 | 8.4 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.8 | 2.3 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.7 | 8.2 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.7 | 8.9 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.7 | 2.9 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.7 | 6.5 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.7 | 4.3 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.7 | 7.2 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.7 | 5.7 | GO:0015677 | copper ion import(GO:0015677) |
| 0.7 | 2.8 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.7 | 2.1 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.7 | 6.0 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.7 | 4.0 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.7 | 1.3 | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) |
| 0.7 | 6.5 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.6 | 10.4 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.6 | 2.6 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.6 | 6.4 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.6 | 10.9 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.6 | 1.9 | GO:0036233 | glycine import(GO:0036233) |
| 0.6 | 1.2 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.6 | 9.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.6 | 6.1 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.6 | 6.7 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.6 | 1.8 | GO:0042946 | glucoside transport(GO:0042946) glycoside transport(GO:1901656) |
| 0.6 | 4.7 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.6 | 1.8 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.6 | 3.5 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.6 | 7.4 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.6 | 9.0 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.6 | 2.8 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.6 | 3.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.5 | 4.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.5 | 19.0 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.5 | 4.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.5 | 17.2 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.5 | 1.6 | GO:0048633 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.5 | 1.5 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.5 | 0.5 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.5 | 1.5 | GO:0015888 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.5 | 4.9 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.5 | 7.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.5 | 4.3 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.5 | 4.2 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.5 | 1.4 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.5 | 0.5 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
| 0.5 | 1.4 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.5 | 3.6 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.4 | 1.3 | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.4 | 3.1 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.4 | 1.3 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.4 | 6.5 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.4 | 2.6 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.4 | 2.6 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.4 | 1.3 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.4 | 8.6 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.4 | 3.3 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.4 | 11.1 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.4 | 0.8 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.4 | 2.5 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.4 | 2.0 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 0.4 | 1.6 | GO:1902548 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.4 | 3.6 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.4 | 1.6 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.4 | 7.2 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.4 | 3.0 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.4 | 1.9 | GO:0003164 | His-Purkinje system development(GO:0003164) |
| 0.4 | 1.9 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.4 | 7.5 | GO:0046051 | UTP metabolic process(GO:0046051) |
| 0.4 | 7.9 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.4 | 1.5 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.4 | 1.5 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.4 | 2.6 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.4 | 5.7 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.4 | 3.9 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.4 | 2.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.4 | 0.4 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.3 | 1.4 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.3 | 1.4 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.3 | 2.0 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.3 | 4.4 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.3 | 11.5 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.3 | 3.4 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.3 | 1.7 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.3 | 10.5 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.3 | 4.9 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.3 | 1.6 | GO:0051643 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031585) endoplasmic reticulum localization(GO:0051643) |
| 0.3 | 1.3 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.3 | 2.6 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.3 | 2.6 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.3 | 3.6 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.3 | 0.3 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.3 | 3.5 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.3 | 2.2 | GO:0007619 | courtship behavior(GO:0007619) negative regulation of female receptivity(GO:0007621) female courtship behavior(GO:0008050) |
| 0.3 | 0.9 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.3 | 1.5 | GO:0034959 | neuropeptide catabolic process(GO:0010813) substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.3 | 1.2 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.3 | 4.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.3 | 0.9 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.3 | 1.5 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) |
| 0.3 | 1.5 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.3 | 5.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.3 | 4.0 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.3 | 1.4 | GO:0015862 | uridine transport(GO:0015862) |
| 0.3 | 6.2 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.3 | 3.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.3 | 1.7 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.3 | 8.6 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.3 | 0.8 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.3 | 2.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.3 | 5.5 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.3 | 1.9 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.3 | 0.8 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.3 | 10.7 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.3 | 2.9 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.3 | 0.8 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.3 | 1.8 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.3 | 1.5 | GO:0048165 | ovarian cumulus expansion(GO:0001550) oogenesis stage(GO:0022605) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.3 | 7.9 | GO:0019054 | modulation by virus of host process(GO:0019054) |
| 0.3 | 4.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.3 | 10.0 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.2 | 0.7 | GO:1902023 | ornithine transport(GO:0015822) L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) L-ornithine transmembrane transport(GO:1903352) |
| 0.2 | 8.0 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.2 | 3.0 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.2 | 3.4 | GO:0033622 | integrin activation(GO:0033622) |
| 0.2 | 7.6 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.2 | 7.5 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.2 | 1.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.2 | 3.5 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.2 | 3.5 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.2 | 0.7 | GO:0021658 | rhombomere morphogenesis(GO:0021593) rhombomere 3 morphogenesis(GO:0021658) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.2 | 3.0 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.2 | 1.4 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.2 | 2.6 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) |
| 0.2 | 2.6 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.2 | 2.4 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.2 | 4.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.2 | 1.7 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.2 | 4.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 0.6 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.2 | 1.1 | GO:0007440 | foregut morphogenesis(GO:0007440) embryonic foregut morphogenesis(GO:0048617) |
| 0.2 | 4.2 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.2 | 8.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.2 | 4.4 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.2 | 3.4 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.2 | 0.6 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.2 | 6.0 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.2 | 2.8 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.2 | 1.6 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.2 | 0.6 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.2 | 1.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 | 1.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.2 | 1.6 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.2 | 2.4 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.2 | 6.7 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.2 | 0.8 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.2 | 12.0 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.2 | 10.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 3.1 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 4.2 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.2 | 1.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.2 | 1.3 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.2 | 0.5 | GO:0097503 | sialylation(GO:0097503) |
| 0.2 | 10.4 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.2 | 3.6 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.2 | 2.8 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.2 | 1.5 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.2 | 1.7 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.2 | 1.5 | GO:0009048 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.2 | 0.5 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.2 | 1.6 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.2 | 4.4 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.2 | 3.4 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.2 | 0.9 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.2 | 1.5 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.2 | 2.4 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.2 | 5.6 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 1.5 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.7 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 0.6 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 3.5 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 0.3 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.1 | 4.1 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.1 | 0.4 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.1 | 3.6 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.1 | 1.4 | GO:0007379 | segment specification(GO:0007379) |
| 0.1 | 1.1 | GO:0032306 | regulation of prostaglandin secretion(GO:0032306) positive regulation of prostaglandin secretion(GO:0032308) |
| 0.1 | 2.7 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.1 | 0.6 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.1 | 0.1 | GO:0003343 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) positive regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905007) |
| 0.1 | 0.1 | GO:0038027 | apolipoprotein A-I-mediated signaling pathway(GO:0038027) |
| 0.1 | 2.0 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 | 3.6 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.1 | 2.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 0.4 | GO:1902846 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.1 | 1.9 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 | 2.8 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.1 | 5.9 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.1 | 1.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 1.7 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.1 | 0.8 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.1 | 0.3 | GO:0071484 | response to high light intensity(GO:0009644) cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) retinal cell apoptotic process(GO:1990009) |
| 0.1 | 1.1 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.1 | 6.7 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.1 | 0.9 | GO:0035411 | catenin import into nucleus(GO:0035411) |
| 0.1 | 3.8 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 2.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.8 | GO:0030421 | defecation(GO:0030421) |
| 0.1 | 2.3 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 2.3 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 2.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 3.9 | GO:0035924 | cellular response to vascular endothelial growth factor stimulus(GO:0035924) |
| 0.1 | 12.1 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 | 1.6 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.1 | 10.3 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 1.1 | GO:0060148 | positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.1 | 5.4 | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032435) |
| 0.1 | 12.9 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.1 | 0.4 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 | 1.2 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.1 | 2.1 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 0.8 | GO:0072178 | nephric duct development(GO:0072176) nephric duct morphogenesis(GO:0072178) |
| 0.1 | 4.1 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.1 | 4.2 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 3.0 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.1 | 0.2 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 1.2 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.1 | 4.2 | GO:0032760 | positive regulation of tumor necrosis factor production(GO:0032760) |
| 0.1 | 2.0 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 0.4 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.1 | 0.4 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.9 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.1 | 1.2 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 2.2 | GO:0042438 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.1 | 4.5 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.1 | 1.9 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.1 | 0.8 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 0.5 | GO:0060147 | regulation of posttranscriptional gene silencing(GO:0060147) regulation of gene silencing by miRNA(GO:0060964) regulation of gene silencing by RNA(GO:0060966) |
| 0.1 | 1.5 | GO:0050691 | regulation of defense response to virus by host(GO:0050691) |
| 0.1 | 2.4 | GO:0031663 | lipopolysaccharide-mediated signaling pathway(GO:0031663) |
| 0.1 | 1.0 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.4 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 3.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 1.9 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 1.4 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.1 | 2.3 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.1 | 2.0 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.1 | 2.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 2.7 | GO:0018196 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 3.6 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.1 | 1.4 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 | 0.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.2 | GO:0033306 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.1 | 3.9 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 0.9 | GO:0051788 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.1 | 0.3 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.1 | 0.2 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 1.0 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.1 | 0.5 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.1 | 8.2 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.1 | 2.9 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 1.8 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 3.8 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 1.4 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.1 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.7 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.9 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.0 | 11.3 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 0.6 | GO:0046320 | regulation of fatty acid oxidation(GO:0046320) |
| 0.0 | 0.1 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 2.8 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.2 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.2 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 1.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.5 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 1.1 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.0 | 0.3 | GO:1901798 | positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 1.6 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 3.5 | GO:0042594 | response to starvation(GO:0042594) |
| 0.0 | 6.5 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.7 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 1.9 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 1.4 | GO:0007077 | mitotic nuclear envelope disassembly(GO:0007077) |
| 0.0 | 0.6 | GO:0035136 | forelimb morphogenesis(GO:0035136) |
| 0.0 | 1.1 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 2.2 | GO:0006109 | regulation of carbohydrate metabolic process(GO:0006109) |
| 0.0 | 1.3 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 1.1 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.9 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.1 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.5 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.1 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.0 | 0.8 | GO:0098760 | interleukin-7-mediated signaling pathway(GO:0038111) response to interleukin-7(GO:0098760) cellular response to interleukin-7(GO:0098761) |
| 0.0 | 2.2 | GO:0006364 | rRNA processing(GO:0006364) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 16.8 | GO:0034677 | integrin alpha7-beta1 complex(GO:0034677) |
| 4.3 | 12.8 | GO:0005606 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 4.0 | 12.0 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 2.7 | 11.0 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 2.4 | 12.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 2.2 | 6.5 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 1.9 | 11.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 1.8 | 7.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 1.5 | 8.9 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 1.2 | 11.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 1.2 | 8.6 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 1.1 | 4.6 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 1.1 | 2.3 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 1.0 | 13.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 1.0 | 10.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 1.0 | 7.8 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 1.0 | 8.7 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.9 | 6.5 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.9 | 14.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.9 | 6.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.8 | 4.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.8 | 5.0 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.8 | 9.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.7 | 12.6 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.7 | 9.6 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.7 | 2.9 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 0.7 | 3.5 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.7 | 2.8 | GO:0035363 | histone locus body(GO:0035363) |
| 0.6 | 6.1 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.6 | 8.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.5 | 2.7 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.5 | 4.8 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.5 | 17.1 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.5 | 6.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.5 | 4.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.5 | 7.5 | GO:0033179 | proton-transporting two-sector ATPase complex, proton-transporting domain(GO:0033177) proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.4 | 1.3 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.4 | 3.4 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.4 | 2.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.4 | 3.8 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.4 | 9.2 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.4 | 3.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.4 | 11.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.4 | 1.1 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.4 | 1.8 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.4 | 4.0 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.4 | 9.9 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.4 | 3.6 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.3 | 14.0 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.3 | 4.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.3 | 3.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.3 | 2.9 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.3 | 1.0 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.3 | 4.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.3 | 3.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.3 | 6.1 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.3 | 1.7 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.3 | 2.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.3 | 4.8 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.3 | 12.9 | GO:0002102 | podosome(GO:0002102) |
| 0.3 | 7.1 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.3 | 2.7 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.3 | 1.6 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.3 | 8.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.3 | 47.3 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.3 | 3.4 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.3 | 49.9 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.2 | 1.5 | GO:0032279 | axonemal microtubule(GO:0005879) asymmetric synapse(GO:0032279) |
| 0.2 | 4.2 | GO:0036452 | ESCRT complex(GO:0036452) |
| 0.2 | 8.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.2 | 4.6 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.2 | 11.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.2 | 3.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 18.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 1.3 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 0.8 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.2 | 0.6 | GO:0032116 | SMC loading complex(GO:0032116) |
| 0.2 | 1.2 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 1.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 5.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.2 | 0.7 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.2 | 2.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.2 | 5.0 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.2 | 8.7 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.2 | 2.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 1.2 | GO:0032059 | bleb(GO:0032059) |
| 0.2 | 24.6 | GO:0016605 | PML body(GO:0016605) |
| 0.2 | 21.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.2 | 3.8 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.2 | 1.5 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.2 | 28.8 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.2 | 1.6 | GO:0090543 | Flemming body(GO:0090543) |
| 0.2 | 2.6 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.2 | 1.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.2 | 3.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 4.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 7.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 2.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 26.3 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 1.5 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 6.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 7.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 15.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 1.6 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 1.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 2.0 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 1.4 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 0.7 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 20.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 3.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 5.2 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 5.0 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.1 | 3.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 5.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 5.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 2.8 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 5.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 0.7 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 0.6 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 1.0 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) Lewy body(GO:0097413) |
| 0.1 | 34.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 1.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 7.6 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.1 | 5.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 6.1 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 22.2 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 3.7 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 0.4 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 1.4 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 3.2 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 0.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 1.1 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 0.9 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 2.4 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 2.1 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.1 | 0.5 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 3.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 1.6 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 7.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 6.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 13.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.8 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 5.7 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 16.8 | GO:0019866 | organelle inner membrane(GO:0019866) |
| 0.0 | 1.8 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 2.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.5 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.8 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 3.5 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 1.7 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 1.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 9.0 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 2.3 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 1.1 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 11.2 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.0 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 1.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.3 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.4 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 1.6 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 0.7 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.3 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 17.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.7 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 26.6 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 0.9 | GO:0005840 | ribosome(GO:0005840) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 23.0 | GO:1990254 | keratin filament binding(GO:1990254) |
| 3.7 | 11.2 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 3.2 | 9.6 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 3.1 | 9.2 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 2.9 | 11.7 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 2.8 | 13.8 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 2.6 | 13.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 2.6 | 15.8 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 2.6 | 10.2 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 2.5 | 9.9 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 2.4 | 9.7 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 2.1 | 8.4 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 2.1 | 6.3 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 1.8 | 10.8 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 1.7 | 17.0 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 1.6 | 11.0 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 1.5 | 4.6 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 1.5 | 6.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 1.5 | 4.4 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 1.4 | 21.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 1.3 | 8.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 1.3 | 3.8 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 1.2 | 3.6 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 1.2 | 3.5 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 1.2 | 3.5 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 1.2 | 19.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 1.1 | 3.4 | GO:0055100 | adiponectin binding(GO:0055100) |
| 1.1 | 3.4 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 1.1 | 10.6 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 1.0 | 4.2 | GO:0016972 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 1.0 | 3.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 1.0 | 9.7 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 1.0 | 12.6 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 1.0 | 6.7 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 1.0 | 4.8 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.9 | 9.3 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.9 | 2.6 | GO:0080101 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.9 | 2.6 | GO:0031753 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.9 | 2.6 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.8 | 4.9 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.8 | 5.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.8 | 2.4 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.8 | 2.4 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.8 | 3.1 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.8 | 2.3 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.8 | 20.4 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.7 | 5.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.7 | 3.6 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.7 | 6.3 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.7 | 8.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.7 | 4.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.7 | 16.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.7 | 2.0 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.7 | 3.4 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.7 | 2.0 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.7 | 2.6 | GO:0043273 | CTPase activity(GO:0043273) |
| 0.6 | 1.9 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.6 | 8.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.6 | 11.8 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.6 | 8.6 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.6 | 8.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.6 | 1.8 | GO:0042947 | glucoside transmembrane transporter activity(GO:0042947) |
| 0.6 | 12.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.6 | 7.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.6 | 5.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.6 | 10.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.6 | 6.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.5 | 10.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.5 | 12.5 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.5 | 2.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.5 | 19.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.5 | 2.6 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.5 | 8.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.5 | 1.5 | GO:0015563 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) uptake transmembrane transporter activity(GO:0015563) |
| 0.5 | 12.9 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.5 | 3.4 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.5 | 7.7 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.5 | 1.9 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.5 | 5.7 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.5 | 1.8 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.4 | 4.8 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.4 | 6.8 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.4 | 3.0 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.4 | 9.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.4 | 6.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.4 | 1.6 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.4 | 12.4 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.4 | 2.3 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.4 | 5.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.4 | 1.5 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.4 | 11.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.4 | 4.0 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.4 | 0.7 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.4 | 4.3 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.4 | 2.8 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.4 | 2.8 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.3 | 2.4 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.3 | 2.4 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.3 | 2.7 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.3 | 2.0 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.3 | 1.3 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.3 | 1.6 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.3 | 2.9 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.3 | 0.9 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.3 | 4.9 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.3 | 9.1 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.3 | 4.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.3 | 2.7 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.3 | 2.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.3 | 5.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.3 | 3.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.3 | 7.5 | GO:0046961 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.3 | 4.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.3 | 5.9 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.3 | 2.5 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.3 | 1.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.3 | 1.7 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.3 | 4.4 | GO:0048185 | activin binding(GO:0048185) |
| 0.3 | 7.1 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.3 | 0.8 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.3 | 4.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 17.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 0.7 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.2 | 1.2 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.2 | 2.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 1.7 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.2 | 0.7 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 1.2 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.2 | 4.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 1.9 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.2 | 1.7 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 0.9 | GO:0004360 | glutamine-fructose-6-phosphate transaminase (isomerizing) activity(GO:0004360) |
| 0.2 | 1.4 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.2 | 9.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 2.8 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.2 | 1.8 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.2 | 21.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 14.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.2 | 7.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.2 | 0.6 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) |
| 0.2 | 4.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 0.6 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.2 | 8.4 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.2 | 7.0 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 2.7 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 9.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.2 | 23.7 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.2 | 6.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.2 | 5.8 | GO:0071949 | FAD binding(GO:0071949) |
| 0.2 | 1.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 4.3 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.2 | 1.1 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.2 | 5.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 3.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.2 | 4.6 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.2 | 2.9 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.2 | 7.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.2 | 8.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.2 | 2.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.2 | 3.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 5.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 6.0 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.2 | 2.5 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.2 | 0.2 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.2 | 6.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.2 | 2.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 1.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 2.4 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 0.4 | GO:0016422 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 2.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 2.0 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 14.9 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 1.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 15.6 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.1 | 2.5 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 4.1 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 2.4 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 2.0 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 1.5 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.7 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.5 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 2.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 3.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.8 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 6.4 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 27.8 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.1 | 0.9 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 2.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 28.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 5.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.1 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 4.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 4.9 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 3.0 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 14.3 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 1.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 2.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 5.2 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 1.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 8.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 2.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.4 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 1.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 2.1 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.1 | 2.3 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 1.6 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 5.1 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.1 | 0.6 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 1.3 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.1 | 0.6 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 1.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.2 | GO:0019166 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 1.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 25.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 0.2 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.1 | 1.4 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 0.8 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 2.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.3 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.1 | 3.8 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 0.8 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 9.5 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 1.9 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.1 | 1.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 5.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.4 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 1.9 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 0.4 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.4 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 5.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 17.0 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 5.6 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 2.6 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 1.0 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 2.8 | GO:0008135 | translation factor activity, RNA binding(GO:0008135) |
| 0.0 | 1.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 2.8 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 1.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 5.7 | GO:0061659 | ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.7 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 3.9 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.3 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 1.6 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.9 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.7 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.6 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.0 | 3.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.3 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.2 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 0.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 3.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 37.7 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.7 | 12.8 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.7 | 24.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.6 | 25.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.5 | 7.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.5 | 24.3 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.5 | 6.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.4 | 25.9 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.4 | 9.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.4 | 1.7 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.4 | 24.6 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.4 | 22.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.4 | 7.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.4 | 41.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.4 | 18.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.3 | 13.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.3 | 10.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.3 | 12.2 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.3 | 14.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.3 | 14.9 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.3 | 9.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.3 | 47.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 7.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.2 | 7.8 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.2 | 11.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 13.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 5.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.2 | 10.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 7.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 2.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.2 | 10.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 5.8 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.2 | 3.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 0.9 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 6.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 5.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 2.0 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 4.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 10.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 9.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 7.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 1.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 1.3 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 1.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 4.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 8.1 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 3.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 3.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 24.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 2.3 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 8.4 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 2.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 4.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 3.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 6.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 1.1 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 2.0 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 1.3 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 5.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 1.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 9.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.6 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 1.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 3.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.9 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.8 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.7 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 18.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 1.0 | 11.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 1.0 | 16.8 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.9 | 31.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.7 | 12.7 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.7 | 13.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.7 | 10.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.6 | 19.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.6 | 25.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.6 | 13.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.6 | 3.9 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.5 | 13.2 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.5 | 17.9 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.5 | 15.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.5 | 13.4 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.5 | 4.9 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.5 | 3.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.5 | 11.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.5 | 7.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.4 | 5.3 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.4 | 9.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.4 | 3.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.4 | 8.0 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.4 | 3.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.4 | 12.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.4 | 7.0 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.4 | 6.6 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.4 | 5.8 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.3 | 18.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.3 | 7.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 4.0 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.3 | 11.2 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.3 | 13.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.3 | 9.1 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.3 | 13.5 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.3 | 4.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.3 | 2.3 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.3 | 6.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.3 | 0.5 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.3 | 1.6 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.2 | 1.9 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.2 | 10.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.2 | 8.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.2 | 4.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 6.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.2 | 7.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 14.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.2 | 3.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.2 | 3.5 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.2 | 3.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 10.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 1.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.2 | 5.2 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 1.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 1.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.4 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 1.8 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 2.8 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 15.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 4.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 2.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.0 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 4.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 3.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 4.9 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 2.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.6 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 2.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 9.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 2.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 2.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 3.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 0.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 4.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 4.0 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 6.2 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 3.0 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 1.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 2.4 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.1 | 1.0 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.1 | 1.8 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.7 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 2.4 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.1 | 0.5 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.9 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 1.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.8 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.1 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 1.4 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 1.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.4 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |