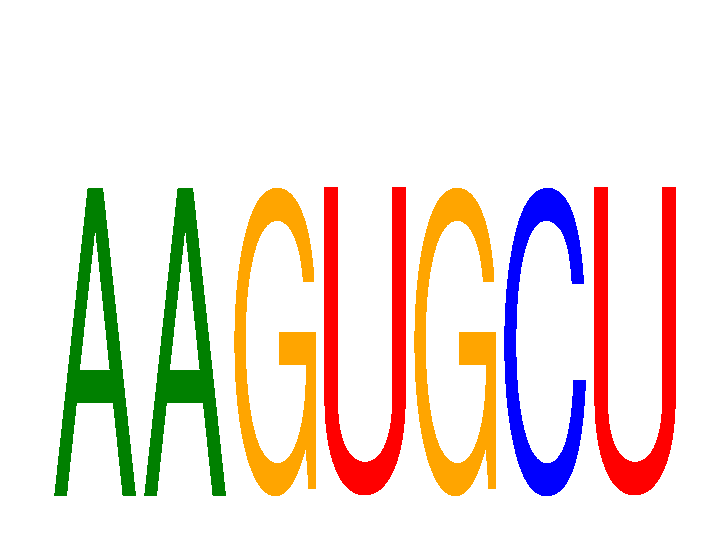Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for AAGUGCU
Z-value: 0.43
miRNA associated with seed AAGUGCU
| Name | miRBASE accession |
|---|---|
|
hsa-miR-302a-3p
|
MIMAT0000684 |
|
hsa-miR-302b-3p
|
MIMAT0000715 |
|
hsa-miR-302c-3p.1
|
MIMAT0000717 |
|
hsa-miR-302d-3p
|
MIMAT0000718 |
|
hsa-miR-302e
|
MIMAT0005931 |
|
hsa-miR-372-3p
|
MIMAT0000724 |
|
hsa-miR-373-3p
|
MIMAT0000726 |
|
hsa-miR-520a-3p
|
MIMAT0002834 |
|
hsa-miR-520b
|
MIMAT0002843 |
|
hsa-miR-520c-3p
|
MIMAT0002846 |
|
hsa-miR-520d-3p
|
MIMAT0002856 |
|
hsa-miR-520e
|
MIMAT0002825 |
Activity profile of AAGUGCU motif
Sorted Z-values of AAGUGCU motif
Network of associatons between targets according to the STRING database.
First level regulatory network of AAGUGCU
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_66613208 | 5.24 |
ENST00000522677.8
|
MYBL1
|
MYB proto-oncogene like 1 |
| chr12_-_44875647 | 4.33 |
ENST00000395487.6
|
NELL2
|
neural EGFL like 2 |
| chr11_+_118530990 | 4.01 |
ENST00000411589.6
ENST00000359862.8 ENST00000442938.6 |
TMEM25
|
transmembrane protein 25 |
| chr6_+_11537738 | 3.83 |
ENST00000379426.2
|
TMEM170B
|
transmembrane protein 170B |
| chr14_-_99272184 | 3.60 |
ENST00000357195.8
|
BCL11B
|
BAF chromatin remodeling complex subunit BCL11B |
| chr16_+_19113955 | 3.12 |
ENST00000381440.5
ENST00000564808.6 ENST00000568526.1 |
ITPRIPL2
ENSG00000261427.6
|
ITPRIP like 2 novel transcript |
| chr8_+_42896883 | 3.00 |
ENST00000307602.9
|
HOOK3
|
hook microtubule tethering protein 3 |
| chrX_+_16719595 | 2.87 |
ENST00000380155.4
|
SYAP1
|
synapse associated protein 1 |
| chr19_+_16197900 | 2.85 |
ENST00000429941.6
ENST00000291439.8 ENST00000444449.6 ENST00000589822.5 |
AP1M1
|
adaptor related protein complex 1 subunit mu 1 |
| chr10_-_27240743 | 2.83 |
ENST00000677901.1
ENST00000677960.1 ENST00000677440.1 ENST00000396271.8 ENST00000677141.1 ENST00000677311.1 ENST00000677667.1 ENST00000677200.1 ENST00000676997.1 ENST00000676511.1 |
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr14_-_59630582 | 2.68 |
ENST00000395090.5
|
RTN1
|
reticulon 1 |
| chr3_+_43286512 | 2.53 |
ENST00000454177.5
ENST00000429705.6 ENST00000296088.12 ENST00000437827.1 |
SNRK
|
SNF related kinase |
| chr4_-_108168919 | 2.48 |
ENST00000265165.6
|
LEF1
|
lymphoid enhancer binding factor 1 |
| chr5_+_72107453 | 2.40 |
ENST00000296755.12
ENST00000511641.2 |
MAP1B
|
microtubule associated protein 1B |
| chr2_-_96145431 | 2.18 |
ENST00000288943.5
|
DUSP2
|
dual specificity phosphatase 2 |
| chr8_-_81112055 | 2.03 |
ENST00000220597.4
|
PAG1
|
phosphoprotein membrane anchor with glycosphingolipid microdomains 1 |
| chr5_-_44389407 | 2.01 |
ENST00000264664.5
|
FGF10
|
fibroblast growth factor 10 |
| chr4_-_82798735 | 1.98 |
ENST00000273908.4
ENST00000319540.9 |
SCD5
|
stearoyl-CoA desaturase 5 |
| chr17_-_42745025 | 1.91 |
ENST00000592492.5
ENST00000585893.5 ENST00000593214.5 ENST00000590078.5 ENST00000428826.7 ENST00000586382.5 ENST00000415827.6 ENST00000592743.5 ENST00000586089.5 |
EZH1
|
enhancer of zeste 1 polycomb repressive complex 2 subunit |
| chr6_+_125790922 | 1.88 |
ENST00000453302.5
ENST00000417494.5 ENST00000392477.7 ENST00000229634.13 |
NCOA7
|
nuclear receptor coactivator 7 |
| chr3_-_50567646 | 1.87 |
ENST00000426034.5
ENST00000441239.5 |
C3orf18
|
chromosome 3 open reading frame 18 |
| chr22_+_29073024 | 1.85 |
ENST00000400335.9
|
KREMEN1
|
kringle containing transmembrane protein 1 |
| chr12_+_119593758 | 1.82 |
ENST00000426426.3
|
TMEM233
|
transmembrane protein 233 |
| chr6_+_41072939 | 1.80 |
ENST00000341376.11
ENST00000353205.5 |
NFYA
|
nuclear transcription factor Y subunit alpha |
| chr10_-_118754956 | 1.78 |
ENST00000369151.8
|
CACUL1
|
CDK2 associated cullin domain 1 |
| chr1_+_226223618 | 1.78 |
ENST00000542034.5
ENST00000366810.6 |
MIXL1
|
Mix paired-like homeobox |
| chrX_+_111096136 | 1.77 |
ENST00000372007.10
|
PAK3
|
p21 (RAC1) activated kinase 3 |
| chr6_-_154356735 | 1.75 |
ENST00000367220.8
ENST00000265198.8 ENST00000520261.1 |
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr17_+_7281711 | 1.71 |
ENST00000317370.13
ENST00000571308.5 |
SLC2A4
|
solute carrier family 2 member 4 |
| chr9_-_127980976 | 1.68 |
ENST00000373095.6
|
FAM102A
|
family with sequence similarity 102 member A |
| chr11_-_132943671 | 1.67 |
ENST00000331898.11
|
OPCML
|
opioid binding protein/cell adhesion molecule like |
| chr1_-_235328147 | 1.66 |
ENST00000264183.9
ENST00000418304.1 ENST00000349213.7 |
ARID4B
|
AT-rich interaction domain 4B |
| chrX_+_23334841 | 1.64 |
ENST00000379361.5
|
PTCHD1
|
patched domain containing 1 |
| chr17_-_50130121 | 1.64 |
ENST00000330175.9
|
SAMD14
|
sterile alpha motif domain containing 14 |
| chr7_-_20217342 | 1.62 |
ENST00000400331.10
ENST00000332878.8 |
MACC1
|
MET transcriptional regulator MACC1 |
| chr3_-_12967668 | 1.61 |
ENST00000273221.8
|
IQSEC1
|
IQ motif and Sec7 domain ArfGEF 1 |
| chr9_-_83956677 | 1.61 |
ENST00000376344.8
|
C9orf64
|
chromosome 9 open reading frame 64 |
| chr16_-_53503192 | 1.59 |
ENST00000568596.5
ENST00000394657.12 ENST00000570004.5 ENST00000564497.1 ENST00000300245.8 |
AKTIP
|
AKT interacting protein |
| chr20_+_58309704 | 1.55 |
ENST00000244040.4
|
RAB22A
|
RAB22A, member RAS oncogene family |
| chr2_-_234497035 | 1.52 |
ENST00000390645.2
ENST00000339728.6 |
ARL4C
|
ADP ribosylation factor like GTPase 4C |
| chr1_-_225889143 | 1.51 |
ENST00000272134.5
|
LEFTY1
|
left-right determination factor 1 |
| chr14_-_44961889 | 1.49 |
ENST00000579157.1
ENST00000396128.9 ENST00000556500.1 |
KLHL28
|
kelch like family member 28 |
| chr21_-_31558977 | 1.48 |
ENST00000286827.7
ENST00000541036.5 |
TIAM1
|
TIAM Rac1 associated GEF 1 |
| chr22_+_39994926 | 1.48 |
ENST00000333407.11
|
FAM83F
|
family with sequence similarity 83 member F |
| chr20_-_4823597 | 1.44 |
ENST00000379400.8
|
RASSF2
|
Ras association domain family member 2 |
| chr11_+_118436464 | 1.42 |
ENST00000389506.10
ENST00000534358.8 ENST00000531904.6 ENST00000649699.1 |
KMT2A
|
lysine methyltransferase 2A |
| chr7_-_44885446 | 1.36 |
ENST00000395699.5
|
PURB
|
purine rich element binding protein B |
| chr6_+_87155537 | 1.36 |
ENST00000369577.8
ENST00000518845.1 ENST00000339907.8 ENST00000496806.2 |
ZNF292
|
zinc finger protein 292 |
| chr3_+_20040437 | 1.34 |
ENST00000263754.5
|
KAT2B
|
lysine acetyltransferase 2B |
| chr1_-_24964984 | 1.33 |
ENST00000338888.3
ENST00000399916.5 |
RUNX3
|
RUNX family transcription factor 3 |
| chr15_+_68578970 | 1.33 |
ENST00000261861.10
|
CORO2B
|
coronin 2B |
| chr12_+_4909895 | 1.33 |
ENST00000638821.1
ENST00000382545.5 |
ENSG00000256654.4
KCNA1
|
novel transcript, sense overlapping KCNA1 potassium voltage-gated channel subfamily A member 1 |
| chr20_-_49278034 | 1.32 |
ENST00000371744.5
ENST00000396105.6 ENST00000371752.5 |
ZNFX1
|
zinc finger NFX1-type containing 1 |
| chr12_+_67648737 | 1.31 |
ENST00000344096.4
ENST00000393555.3 |
DYRK2
|
dual specificity tyrosine phosphorylation regulated kinase 2 |
| chr7_+_77696423 | 1.31 |
ENST00000334955.13
|
RSBN1L
|
round spermatid basic protein 1 like |
| chr6_-_142946312 | 1.27 |
ENST00000367604.6
|
HIVEP2
|
HIVEP zinc finger 2 |
| chr1_-_230426293 | 1.27 |
ENST00000391860.7
|
PGBD5
|
piggyBac transposable element derived 5 |
| chr5_-_115180037 | 1.26 |
ENST00000514154.1
ENST00000282369.7 |
TRIM36
|
tripartite motif containing 36 |
| chr1_+_244051275 | 1.22 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr2_+_197804583 | 1.19 |
ENST00000428675.6
|
PLCL1
|
phospholipase C like 1 (inactive) |
| chr2_+_29115367 | 1.19 |
ENST00000320081.10
|
CLIP4
|
CAP-Gly domain containing linker protein family member 4 |
| chr9_-_109167159 | 1.18 |
ENST00000561981.5
|
FRRS1L
|
ferric chelate reductase 1 like |
| chr2_+_69829630 | 1.17 |
ENST00000282570.4
|
GMCL1
|
germ cell-less 1, spermatogenesis associated |
| chr1_+_84078043 | 1.17 |
ENST00000370689.6
ENST00000370688.7 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr3_+_179347686 | 1.16 |
ENST00000471841.6
|
MFN1
|
mitofusin 1 |
| chr6_-_32128191 | 1.16 |
ENST00000453203.2
ENST00000375203.8 ENST00000375201.8 |
ATF6B
|
activating transcription factor 6 beta |
| chr2_+_26848093 | 1.15 |
ENST00000288699.11
|
DPYSL5
|
dihydropyrimidinase like 5 |
| chr6_-_89118002 | 1.15 |
ENST00000452027.3
|
SRSF12
|
serine and arginine rich splicing factor 12 |
| chr1_-_20486197 | 1.14 |
ENST00000375078.4
|
CAMK2N1
|
calcium/calmodulin dependent protein kinase II inhibitor 1 |
| chr15_-_61229297 | 1.13 |
ENST00000335670.11
|
RORA
|
RAR related orphan receptor A |
| chr2_-_25878445 | 1.13 |
ENST00000336112.9
ENST00000435504.9 |
ASXL2
|
ASXL transcriptional regulator 2 |
| chr11_+_129375841 | 1.13 |
ENST00000281437.6
|
BARX2
|
BARX homeobox 2 |
| chr12_+_72272360 | 1.12 |
ENST00000547300.2
ENST00000261180.10 |
TRHDE
|
thyrotropin releasing hormone degrading enzyme |
| chr4_+_44678412 | 1.12 |
ENST00000281543.6
|
GUF1
|
GTP binding elongation factor GUF1 |
| chr4_+_85475131 | 1.10 |
ENST00000395184.6
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr17_-_5234801 | 1.10 |
ENST00000571800.5
ENST00000574081.6 ENST00000399600.8 ENST00000574297.1 |
SCIMP
|
SLP adaptor and CSK interacting membrane protein |
| chr13_+_51584435 | 1.09 |
ENST00000612477.1
ENST00000298125.7 |
WDFY2
|
WD repeat and FYVE domain containing 2 |
| chr10_+_124801799 | 1.07 |
ENST00000298492.6
|
ABRAXAS2
|
abraxas 2, BRISC complex subunit |
| chr9_-_23821275 | 1.07 |
ENST00000380110.8
|
ELAVL2
|
ELAV like RNA binding protein 2 |
| chr16_-_2135898 | 1.06 |
ENST00000262304.9
ENST00000423118.5 |
PKD1
|
polycystin 1, transient receptor potential channel interacting |
| chr9_+_79571767 | 1.06 |
ENST00000376544.7
|
TLE4
|
TLE family member 4, transcriptional corepressor |
| chr8_-_19013693 | 1.06 |
ENST00000327040.13
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr14_-_92040027 | 1.06 |
ENST00000267622.8
|
TRIP11
|
thyroid hormone receptor interactor 11 |
| chr7_-_5781594 | 1.05 |
ENST00000416985.5
ENST00000389902.8 ENST00000425013.6 |
RNF216
|
ring finger protein 216 |
| chr1_-_226737277 | 1.04 |
ENST00000272117.7
|
ITPKB
|
inositol-trisphosphate 3-kinase B |
| chr1_-_207051202 | 1.03 |
ENST00000315927.9
|
YOD1
|
YOD1 deubiquitinase |
| chr8_+_28494190 | 1.02 |
ENST00000537916.2
ENST00000240093.8 ENST00000523546.1 |
FZD3
|
frizzled class receptor 3 |
| chr9_-_16870662 | 1.01 |
ENST00000380672.9
|
BNC2
|
basonuclin 2 |
| chr10_+_61901678 | 1.00 |
ENST00000644638.1
ENST00000681100.1 ENST00000279873.12 |
ARID5B
|
AT-rich interaction domain 5B |
| chr15_+_31326807 | 0.99 |
ENST00000307145.4
|
KLF13
|
Kruppel like factor 13 |
| chr7_-_32891744 | 0.98 |
ENST00000304056.9
|
KBTBD2
|
kelch repeat and BTB domain containing 2 |
| chr1_+_3690654 | 0.98 |
ENST00000378285.5
ENST00000378280.5 ENST00000378288.8 |
TP73
|
tumor protein p73 |
| chr7_+_139341311 | 0.95 |
ENST00000297534.7
ENST00000541515.3 |
FMC1
FMC1-LUC7L2
|
formation of mitochondrial complex V assembly factor 1 homolog FMC1-LUC7L2 readthrough |
| chr7_+_139359846 | 0.95 |
ENST00000619796.4
ENST00000354926.9 |
LUC7L2
|
LUC7 like 2, pre-mRNA splicing factor |
| chr15_-_83284645 | 0.92 |
ENST00000345382.7
|
BNC1
|
basonuclin 1 |
| chr3_-_18425295 | 0.92 |
ENST00000338745.11
ENST00000450898.1 |
SATB1
|
SATB homeobox 1 |
| chr15_-_78234513 | 0.92 |
ENST00000558130.1
ENST00000258873.9 |
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr12_+_8032692 | 0.91 |
ENST00000162391.8
|
FOXJ2
|
forkhead box J2 |
| chr1_+_209827964 | 0.90 |
ENST00000491415.7
|
UTP25
|
UTP25 small subunit processor component |
| chr20_+_62143729 | 0.89 |
ENST00000331758.8
ENST00000450482.5 |
SS18L1
|
SS18L1 subunit of BAF chromatin remodeling complex |
| chr1_-_225941383 | 0.88 |
ENST00000420304.6
|
LEFTY2
|
left-right determination factor 2 |
| chr4_+_74308463 | 0.88 |
ENST00000413830.6
|
EPGN
|
epithelial mitogen |
| chr9_-_19102887 | 0.87 |
ENST00000380502.8
|
HAUS6
|
HAUS augmin like complex subunit 6 |
| chr12_-_56189548 | 0.87 |
ENST00000347471.8
ENST00000267064.8 ENST00000394023.7 |
SMARCC2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily c member 2 |
| chr1_-_113812448 | 0.86 |
ENST00000612242.4
ENST00000261441.9 |
RSBN1
|
round spermatid basic protein 1 |
| chr12_-_16608183 | 0.86 |
ENST00000354662.5
ENST00000538051.5 |
LMO3
|
LIM domain only 3 |
| chr10_+_135067 | 0.85 |
ENST00000381591.5
|
ZMYND11
|
zinc finger MYND-type containing 11 |
| chr8_-_52714414 | 0.84 |
ENST00000435644.6
ENST00000518710.5 ENST00000025008.10 ENST00000517963.1 |
RB1CC1
|
RB1 inducible coiled-coil 1 |
| chr1_-_55215345 | 0.84 |
ENST00000294383.7
|
USP24
|
ubiquitin specific peptidase 24 |
| chr3_-_56801939 | 0.84 |
ENST00000296315.8
ENST00000495373.5 |
ARHGEF3
|
Rho guanine nucleotide exchange factor 3 |
| chr1_-_47231715 | 0.83 |
ENST00000371884.6
|
TAL1
|
TAL bHLH transcription factor 1, erythroid differentiation factor |
| chr8_-_42051978 | 0.82 |
ENST00000265713.8
ENST00000648335.1 ENST00000485568.5 ENST00000426524.6 ENST00000396930.4 ENST00000406337.6 |
KAT6A
|
lysine acetyltransferase 6A |
| chr5_+_149730260 | 0.81 |
ENST00000360453.8
ENST00000394320.7 ENST00000309241.10 |
PPARGC1B
|
PPARG coactivator 1 beta |
| chr5_-_175444132 | 0.81 |
ENST00000393752.3
|
DRD1
|
dopamine receptor D1 |
| chr17_-_68291116 | 0.80 |
ENST00000327268.8
ENST00000580666.6 |
SLC16A6
|
solute carrier family 16 member 6 |
| chr3_-_197749688 | 0.80 |
ENST00000273582.9
|
RUBCN
|
rubicon autophagy regulator |
| chr1_+_28369705 | 0.79 |
ENST00000373839.8
|
PHACTR4
|
phosphatase and actin regulator 4 |
| chr11_-_73598183 | 0.78 |
ENST00000064778.8
|
FAM168A
|
family with sequence similarity 168 member A |
| chr10_+_72215981 | 0.76 |
ENST00000615507.4
ENST00000621663.4 ENST00000299381.5 |
ANAPC16
|
anaphase promoting complex subunit 16 |
| chr7_+_87345656 | 0.75 |
ENST00000331536.8
ENST00000419147.6 ENST00000412227.6 |
CROT
|
carnitine O-octanoyltransferase |
| chr11_-_77820706 | 0.74 |
ENST00000440064.2
ENST00000528095.5 ENST00000308488.11 |
RSF1
|
remodeling and spacing factor 1 |
| chr21_+_45643694 | 0.74 |
ENST00000400314.5
|
PCBP3
|
poly(rC) binding protein 3 |
| chr7_+_120273129 | 0.73 |
ENST00000331113.9
|
KCND2
|
potassium voltage-gated channel subfamily D member 2 |
| chr7_-_142885737 | 0.73 |
ENST00000359396.9
ENST00000436401.1 |
TRPV6
|
transient receptor potential cation channel subfamily V member 6 |
| chr2_-_181680490 | 0.73 |
ENST00000684145.1
ENST00000295108.4 ENST00000684079.1 ENST00000683430.1 |
CERKL
NEUROD1
|
ceramide kinase like neuronal differentiation 1 |
| chr2_-_69643703 | 0.72 |
ENST00000406297.7
ENST00000409085.9 |
AAK1
|
AP2 associated kinase 1 |
| chr7_+_20330893 | 0.72 |
ENST00000222573.5
|
ITGB8
|
integrin subunit beta 8 |
| chr4_-_52659238 | 0.70 |
ENST00000451218.6
ENST00000441222.8 |
USP46
|
ubiquitin specific peptidase 46 |
| chr12_+_55966821 | 0.70 |
ENST00000553376.5
ENST00000440311.6 ENST00000266970.9 ENST00000354056.4 |
CDK2
|
cyclin dependent kinase 2 |
| chr17_+_49788672 | 0.70 |
ENST00000454930.6
ENST00000259021.9 ENST00000509773.5 ENST00000510819.5 ENST00000424009.6 |
KAT7
|
lysine acetyltransferase 7 |
| chr11_-_62727444 | 0.70 |
ENST00000301785.7
|
HNRNPUL2
|
heterogeneous nuclear ribonucleoprotein U like 2 |
| chr17_-_49764123 | 0.69 |
ENST00000240364.7
ENST00000506156.1 |
FAM117A
|
family with sequence similarity 117 member A |
| chr17_+_44187027 | 0.69 |
ENST00000587989.1
ENST00000590235.5 |
TMUB2
|
transmembrane and ubiquitin like domain containing 2 |
| chr12_-_122526929 | 0.69 |
ENST00000331738.12
ENST00000528279.1 ENST00000344591.8 ENST00000526560.6 |
RSRC2
|
arginine and serine rich coiled-coil 2 |
| chr7_+_65873068 | 0.69 |
ENST00000360768.5
|
VKORC1L1
|
vitamin K epoxide reductase complex subunit 1 like 1 |
| chr18_-_47930630 | 0.67 |
ENST00000262160.11
|
SMAD2
|
SMAD family member 2 |
| chr2_-_196171565 | 0.67 |
ENST00000263955.9
|
STK17B
|
serine/threonine kinase 17b |
| chr18_+_34978244 | 0.66 |
ENST00000436190.6
|
MAPRE2
|
microtubule associated protein RP/EB family member 2 |
| chr9_+_36036899 | 0.65 |
ENST00000377966.4
|
RECK
|
reversion inducing cysteine rich protein with kazal motifs |
| chr14_+_35046238 | 0.65 |
ENST00000280987.9
|
FAM177A1
|
family with sequence similarity 177 member A1 |
| chr11_-_68213277 | 0.65 |
ENST00000401547.6
ENST00000304363.9 ENST00000453170.5 |
KMT5B
|
lysine methyltransferase 5B |
| chrX_+_134373297 | 0.64 |
ENST00000332070.7
ENST00000370803.8 ENST00000625464.2 ENST00000370799.5 |
PHF6
|
PHD finger protein 6 |
| chr2_-_102736819 | 0.64 |
ENST00000258436.10
|
MFSD9
|
major facilitator superfamily domain containing 9 |
| chr2_-_23927107 | 0.64 |
ENST00000238789.10
|
ATAD2B
|
ATPase family AAA domain containing 2B |
| chr12_-_95073580 | 0.63 |
ENST00000393101.7
ENST00000333003.10 ENST00000622476.4 |
NR2C1
|
nuclear receptor subfamily 2 group C member 1 |
| chr17_-_58517835 | 0.63 |
ENST00000579921.1
ENST00000579925.5 ENST00000323456.9 |
MTMR4
|
myotubularin related protein 4 |
| chr1_-_175743529 | 0.63 |
ENST00000367674.7
ENST00000263525.6 |
TNR
|
tenascin R |
| chr9_-_37465402 | 0.63 |
ENST00000307750.5
|
ZBTB5
|
zinc finger and BTB domain containing 5 |
| chr20_-_37095985 | 0.62 |
ENST00000344359.7
ENST00000373664.8 |
RBL1
|
RB transcriptional corepressor like 1 |
| chr10_+_91220603 | 0.62 |
ENST00000336126.6
|
PCGF5
|
polycomb group ring finger 5 |
| chr14_+_52267683 | 0.62 |
ENST00000306051.3
ENST00000553372.1 |
PTGDR
|
prostaglandin D2 receptor |
| chr16_-_4273014 | 0.62 |
ENST00000204517.11
|
TFAP4
|
transcription factor AP-4 |
| chr2_-_73113018 | 0.62 |
ENST00000258098.6
|
RAB11FIP5
|
RAB11 family interacting protein 5 |
| chr11_+_61752603 | 0.61 |
ENST00000278836.10
|
MYRF
|
myelin regulatory factor |
| chr5_+_96936071 | 0.60 |
ENST00000231368.10
|
LNPEP
|
leucyl and cystinyl aminopeptidase |
| chr8_-_73878816 | 0.59 |
ENST00000602593.6
ENST00000651945.1 ENST00000419880.7 ENST00000517608.5 ENST00000650817.1 |
UBE2W
|
ubiquitin conjugating enzyme E2 W |
| chr11_-_108593738 | 0.59 |
ENST00000525344.5
ENST00000265843.9 |
EXPH5
|
exophilin 5 |
| chr13_+_21671067 | 0.59 |
ENST00000382353.6
|
FGF9
|
fibroblast growth factor 9 |
| chr6_+_108166015 | 0.59 |
ENST00000368986.9
|
NR2E1
|
nuclear receptor subfamily 2 group E member 1 |
| chr14_+_56579782 | 0.58 |
ENST00000261556.11
ENST00000538838.5 |
TMEM260
|
transmembrane protein 260 |
| chr3_-_15065240 | 0.58 |
ENST00000449354.6
ENST00000253686.7 ENST00000444840.6 |
MRPS25
|
mitochondrial ribosomal protein S25 |
| chr11_+_33257265 | 0.57 |
ENST00000303296.9
ENST00000379016.7 |
HIPK3
|
homeodomain interacting protein kinase 3 |
| chr11_+_67119245 | 0.57 |
ENST00000529006.7
ENST00000398645.6 |
KDM2A
|
lysine demethylase 2A |
| chr7_+_100015588 | 0.57 |
ENST00000324306.11
ENST00000426572.5 |
ZKSCAN1
|
zinc finger with KRAB and SCAN domains 1 |
| chr3_+_10164883 | 0.56 |
ENST00000256458.5
|
IRAK2
|
interleukin 1 receptor associated kinase 2 |
| chr20_+_32277626 | 0.55 |
ENST00000375712.4
|
KIF3B
|
kinesin family member 3B |
| chr17_+_75721451 | 0.55 |
ENST00000200181.8
|
ITGB4
|
integrin subunit beta 4 |
| chr9_-_135907509 | 0.55 |
ENST00000389532.9
|
CAMSAP1
|
calmodulin regulated spectrin associated protein 1 |
| chr8_-_27772585 | 0.54 |
ENST00000522915.5
ENST00000356537.9 |
CCDC25
|
coiled-coil domain containing 25 |
| chr8_-_56211257 | 0.54 |
ENST00000316981.8
ENST00000423799.6 ENST00000429357.2 |
PLAG1
|
PLAG1 zinc finger |
| chr2_-_239400949 | 0.54 |
ENST00000345617.7
|
HDAC4
|
histone deacetylase 4 |
| chr12_-_57742120 | 0.54 |
ENST00000257897.7
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr9_-_122913299 | 0.52 |
ENST00000373659.4
|
ZBTB6
|
zinc finger and BTB domain containing 6 |
| chr2_+_238426920 | 0.52 |
ENST00000264607.9
|
ASB1
|
ankyrin repeat and SOCS box containing 1 |
| chr1_-_205631962 | 0.52 |
ENST00000289703.8
ENST00000357992.9 |
ELK4
|
ETS transcription factor ELK4 |
| chr3_-_101677119 | 0.52 |
ENST00000312938.5
|
ZBTB11
|
zinc finger and BTB domain containing 11 |
| chr15_-_65517244 | 0.51 |
ENST00000341861.9
|
DPP8
|
dipeptidyl peptidase 8 |
| chr8_-_59119121 | 0.50 |
ENST00000361421.2
|
TOX
|
thymocyte selection associated high mobility group box |
| chr15_-_45187955 | 0.50 |
ENST00000560471.5
ENST00000560540.5 |
SHF
|
Src homology 2 domain containing F |
| chr10_+_100535927 | 0.50 |
ENST00000299163.7
|
HIF1AN
|
hypoxia inducible factor 1 subunit alpha inhibitor |
| chr14_+_103334176 | 0.50 |
ENST00000560338.5
ENST00000560763.5 ENST00000216554.8 |
EIF5
|
eukaryotic translation initiation factor 5 |
| chr8_+_28890365 | 0.49 |
ENST00000519662.5
ENST00000558662.5 ENST00000287701.15 ENST00000523613.5 ENST00000560599.5 ENST00000397358.7 |
HMBOX1
|
homeobox containing 1 |
| chr1_-_84690406 | 0.49 |
ENST00000605755.5
ENST00000342203.8 ENST00000437941.6 |
SSX2IP
|
SSX family member 2 interacting protein |
| chr2_-_73269483 | 0.48 |
ENST00000295133.9
|
FBXO41
|
F-box protein 41 |
| chr2_-_24047348 | 0.47 |
ENST00000406895.3
|
WDCP
|
WD repeat and coiled coil containing |
| chr2_+_15940537 | 0.47 |
ENST00000281043.4
ENST00000638417.1 |
MYCN
|
MYCN proto-oncogene, bHLH transcription factor |
| chr3_+_138187248 | 0.47 |
ENST00000538260.5
ENST00000481646.5 ENST00000491704.5 ENST00000469044.6 ENST00000461600.5 ENST00000466749.5 ENST00000358441.6 ENST00000489213.5 |
ARMC8
|
armadillo repeat containing 8 |
| chr10_+_14878848 | 0.46 |
ENST00000433779.5
ENST00000378325.7 ENST00000354919.11 ENST00000313519.9 ENST00000420416.1 |
SUV39H2
|
suppressor of variegation 3-9 homolog 2 |
| chr3_+_15206179 | 0.45 |
ENST00000253693.7
|
CAPN7
|
calpain 7 |
| chr9_+_126805003 | 0.45 |
ENST00000449886.5
ENST00000450858.1 ENST00000373464.5 |
ZBTB43
|
zinc finger and BTB domain containing 43 |
| chr7_-_152435786 | 0.45 |
ENST00000682283.1
ENST00000679882.1 ENST00000452749.2 ENST00000683616.1 ENST00000262189.11 ENST00000683490.1 ENST00000681082.1 ENST00000684550.1 |
KMT2C
|
lysine methyltransferase 2C |
| chr3_-_171460368 | 0.44 |
ENST00000436636.7
ENST00000465393.1 ENST00000341852.10 |
TNIK
|
TRAF2 and NCK interacting kinase |
| chr13_-_26221703 | 0.44 |
ENST00000381570.7
ENST00000346166.7 |
RNF6
|
ring finger protein 6 |
| chr4_-_73258785 | 0.44 |
ENST00000639793.1
ENST00000358602.9 ENST00000330838.10 ENST00000561029.1 |
ANKRD17
|
ankyrin repeat domain 17 |
| chr9_-_10612966 | 0.44 |
ENST00000381196.9
|
PTPRD
|
protein tyrosine phosphatase receptor type D |
| chr3_-_101320558 | 0.43 |
ENST00000193391.8
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2 |
| chr19_-_4066892 | 0.43 |
ENST00000322357.9
|
ZBTB7A
|
zinc finger and BTB domain containing 7A |
| chr6_-_154510675 | 0.43 |
ENST00000607772.6
|
CNKSR3
|
CNKSR family member 3 |
| chr21_-_34888683 | 0.41 |
ENST00000344691.8
ENST00000358356.9 |
RUNX1
|
RUNX family transcription factor 1 |
| chr10_+_1049476 | 0.41 |
ENST00000358220.5
|
WDR37
|
WD repeat domain 37 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.8 | 2.3 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.7 | 3.7 | GO:0030242 | pexophagy(GO:0030242) |
| 0.6 | 2.3 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.5 | 3.0 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.5 | 2.5 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.5 | 2.5 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.5 | 1.4 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.4 | 1.5 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.3 | 1.0 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.3 | 1.3 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.3 | 4.3 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.3 | 1.4 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.3 | 0.8 | GO:0030221 | basophil differentiation(GO:0030221) positive regulation of chromatin assembly or disassembly(GO:0045799) astrocyte fate commitment(GO:0060018) |
| 0.3 | 1.1 | GO:0072237 | metanephric proximal tubule development(GO:0072237) |
| 0.3 | 0.8 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of autophagosome maturation(GO:1901097) |
| 0.3 | 0.8 | GO:0061386 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.3 | 0.8 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.2 | 1.3 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.2 | 0.8 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.2 | 0.6 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.2 | 1.2 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.2 | 1.7 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.2 | 0.7 | GO:0072708 | response to sorbitol(GO:0072708) |
| 0.2 | 2.0 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.2 | 0.8 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.2 | 0.6 | GO:0048690 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.1 | 1.0 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) |
| 0.1 | 0.4 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.1 | 2.9 | GO:0035646 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.1 | 2.4 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.1 | 0.7 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 1.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.5 | GO:0061428 | peptidyl-aspartic acid hydroxylation(GO:0042264) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 | 0.4 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.1 | 1.5 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 0.4 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.7 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.2 | GO:0030806 | negative regulation of cyclic nucleotide catabolic process(GO:0030806) negative regulation of cAMP catabolic process(GO:0030821) |
| 0.1 | 2.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.2 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.9 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 1.1 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 1.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 1.8 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 1.2 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.1 | 1.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 0.9 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 1.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 0.4 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 1.2 | GO:0030238 | male sex determination(GO:0030238) |
| 0.1 | 0.3 | GO:0003099 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.1 | 1.0 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 1.6 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 1.6 | GO:0060004 | reflex(GO:0060004) |
| 0.1 | 1.9 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.1 | 1.7 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 1.8 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 0.4 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 0.7 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.1 | 0.6 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 0.5 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.1 | 0.6 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.6 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.1 | 1.7 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.1 | 1.7 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.7 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 0.7 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.2 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.1 | 0.2 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.1 | 0.2 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.1 | 2.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 1.4 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.5 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.7 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.9 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.4 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.4 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.2 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.6 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.4 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.6 | GO:0071468 | cellular response to acidic pH(GO:0071468) |
| 0.0 | 0.2 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.7 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.0 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.0 | 0.5 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.3 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.0 | 1.1 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.6 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.1 | GO:2000836 | androgen secretion(GO:0035935) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) |
| 0.0 | 2.2 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.0 | 0.5 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 1.4 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 0.4 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.9 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.8 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 1.3 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 1.5 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 1.3 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 0.3 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.0 | 0.4 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.4 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.8 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.6 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 1.8 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 0.5 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 1.1 | GO:0050691 | regulation of defense response to virus by host(GO:0050691) |
| 0.0 | 1.6 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.7 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.3 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.6 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.4 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.2 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 1.0 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.1 | GO:0044830 | modulation by host of viral RNA genome replication(GO:0044830) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.0 | 0.2 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 1.1 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.2 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.0 | 1.1 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.2 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 2.0 | GO:0050868 | negative regulation of T cell activation(GO:0050868) |
| 0.0 | 0.0 | GO:0002537 | nitric oxide production involved in inflammatory response(GO:0002537) |
| 0.0 | 0.2 | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902237) |
| 0.0 | 0.1 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.0 | 1.2 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 1.4 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.0 | 0.8 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.0 | 0.4 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.4 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.8 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.3 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 1.7 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.0 | 0.3 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.5 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.3 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.6 | GO:0070695 | FHF complex(GO:0070695) |
| 0.4 | 1.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) outer acrosomal membrane(GO:0002081) |
| 0.2 | 0.7 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.2 | 0.7 | GO:0097135 | X chromosome(GO:0000805) cyclin E2-CDK2 complex(GO:0097135) |
| 0.2 | 0.6 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.2 | 1.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 1.8 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.3 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 2.5 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 0.7 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 0.6 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.1 | 0.6 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.1 | 1.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 1.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 1.2 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.1 | 0.9 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.6 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.2 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.1 | 0.4 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 1.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 0.8 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.7 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 0.4 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 1.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.5 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.1 | 1.1 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 1.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.3 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.1 | 0.2 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 2.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.2 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.9 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 1.4 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) interchromatin granule(GO:0035061) |
| 0.0 | 0.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 1.8 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 1.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 3.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.9 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.9 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.9 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 2.3 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.3 | GO:0030681 | multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 1.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.6 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 1.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 3.0 | GO:0031461 | cullin-RING ubiquitin ligase complex(GO:0031461) |
| 0.0 | 1.8 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.7 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 2.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.5 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.2 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.2 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 1.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.5 | 2.0 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.5 | 2.0 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.5 | 1.4 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.5 | 5.0 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.4 | 1.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.3 | 1.0 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.2 | 1.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 0.8 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.2 | 1.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 0.7 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 2.4 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.1 | 1.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 2.8 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 0.4 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 1.7 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.9 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 1.8 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.7 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.1 | 0.5 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.1 | 0.7 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.1 | 0.7 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 0.6 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 1.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 0.7 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.8 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 0.5 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 1.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.3 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.1 | 2.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 2.3 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 0.5 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.5 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.1 | 1.0 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 1.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 1.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.6 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 0.7 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.1 | 0.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 1.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.9 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.9 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.3 | GO:0000406 | double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.1 | 1.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.2 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.1 | 0.6 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.5 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.7 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 1.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 1.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.0 | 0.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 1.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.6 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 4.6 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.7 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.9 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.3 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) |
| 0.0 | 0.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.3 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.7 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.3 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 1.9 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 1.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.4 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.0 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 7.2 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 0.3 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.5 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.8 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 8.6 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) |
| 0.0 | 0.0 | GO:0015181 | L-ornithine transmembrane transporter activity(GO:0000064) arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.1 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.6 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.1 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 1.0 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 2.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 0.6 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 5.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 1.8 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 2.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 2.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 2.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.5 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 2.8 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.5 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 2.0 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.8 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 2.4 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 3.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.4 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 3.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.9 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 2.6 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 3.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.0 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 1.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 0.6 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 0.6 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.6 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 2.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 2.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.2 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 1.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.3 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |