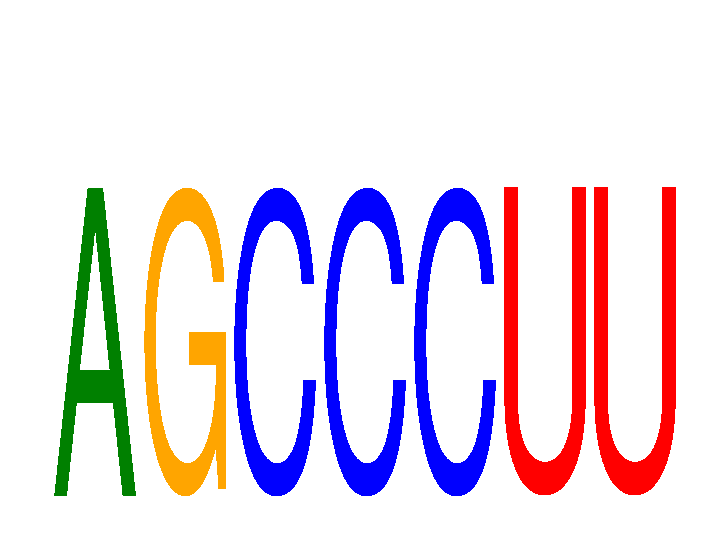Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for AGCCCUU
Z-value: 2.35
miRNA associated with seed AGCCCUU
| Name | miRBASE accession |
|---|---|
|
hsa-miR-129-1-3p
|
MIMAT0004548 |
|
hsa-miR-129-2-3p
|
MIMAT0004605 |
Activity profile of AGCCCUU motif
Sorted Z-values of AGCCCUU motif
Network of associatons between targets according to the STRING database.
First level regulatory network of AGCCCUU
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 11.2 | GO:1902769 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 2.7 | 21.8 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 2.5 | 7.5 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 2.3 | 11.6 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 2.3 | 27.2 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 2.1 | 6.4 | GO:0071206 | establishment of protein localization to juxtaparanode region of axon(GO:0071206) |
| 1.8 | 5.5 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 1.8 | 5.5 | GO:0070350 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 1.5 | 4.6 | GO:0019075 | virus maturation(GO:0019075) |
| 1.3 | 5.4 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 1.3 | 8.8 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 1.2 | 3.7 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 1.1 | 6.8 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 1.1 | 7.4 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 1.0 | 6.0 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.8 | 2.5 | GO:0070650 | endoplasmic reticulum polarization(GO:0061163) actin filament bundle retrograde transport(GO:0061573) actin filament bundle distribution(GO:0070650) |
| 0.7 | 2.2 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.7 | 4.2 | GO:1903059 | regulation of protein lipidation(GO:1903059) positive regulation of protein lipidation(GO:1903061) |
| 0.6 | 3.8 | GO:1903750 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.6 | 1.7 | GO:2000374 | regulation of oxygen metabolic process(GO:2000374) |
| 0.6 | 3.9 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.5 | 1.6 | GO:0036304 | umbilical cord morphogenesis(GO:0036304) umbilical cord development(GO:0061027) |
| 0.4 | 2.0 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.4 | 3.9 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.4 | 19.5 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.4 | 3.4 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.4 | 3.4 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.4 | 5.8 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.4 | 2.2 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.4 | 3.2 | GO:0015886 | heme transport(GO:0015886) |
| 0.3 | 13.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.3 | 4.0 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.3 | 3.8 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.3 | 3.0 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.3 | 1.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.3 | 1.3 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.3 | 4.3 | GO:0038065 | collagen-activated signaling pathway(GO:0038065) |
| 0.2 | 1.0 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.2 | 3.9 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.2 | 6.2 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.2 | 1.8 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.2 | 10.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 0.6 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.2 | 1.0 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.2 | 0.6 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.2 | 3.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.2 | 2.4 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.2 | 0.6 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.2 | 2.4 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.2 | 0.7 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.2 | 0.9 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 1.2 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.2 | 13.8 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.2 | 2.6 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.2 | 3.8 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 2.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 1.2 | GO:1901538 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.1 | 1.7 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 1.5 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.1 | 1.9 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 | 0.9 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 0.8 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.9 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 4.5 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.1 | 1.7 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 0.7 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.1 | 2.8 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 6.4 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 1.0 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.1 | 2.0 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 0.3 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.1 | 0.7 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 2.7 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 2.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 3.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.8 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.2 | GO:0021836 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.0 | 0.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.8 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.9 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 2.3 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.6 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.3 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 4.5 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 1.2 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.2 | GO:0070432 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070432) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.0 | 4.2 | GO:0050868 | negative regulation of T cell activation(GO:0050868) |
| 0.0 | 0.2 | GO:1900107 | regulation of nodal signaling pathway(GO:1900107) |
| 0.0 | 1.0 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.7 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.9 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0019046 | release from viral latency(GO:0019046) regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.1 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.0 | 0.6 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 5.6 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 0.2 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 1.7 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 2.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.4 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.0 | 2.4 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 0.7 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 6.4 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.7 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 | 1.2 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.5 | GO:0051567 | histone H3-K9 methylation(GO:0051567) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 0.1 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 1.0 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 5.5 | GO:0007283 | spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.8 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 1.9 | 11.6 | GO:0045298 | tubulin complex(GO:0045298) |
| 1.5 | 4.5 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 1.2 | 6.0 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 1.2 | 7.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 1.1 | 4.3 | GO:0034681 | integrin alpha11-beta1 complex(GO:0034681) |
| 0.8 | 3.9 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.6 | 11.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.6 | 14.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.6 | 20.5 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.6 | 7.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.5 | 4.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.5 | 6.8 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.4 | 3.9 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.3 | 2.0 | GO:0032437 | muscle tendon junction(GO:0005927) cuticular plate(GO:0032437) |
| 0.3 | 7.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 13.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 1.9 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.2 | 1.9 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.2 | 4.0 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 1.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.2 | 0.6 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.2 | 4.7 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.2 | 11.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 3.8 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 3.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 3.9 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 2.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 29.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 0.7 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 2.2 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.9 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 4.1 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 0.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 5.5 | GO:0031970 | organelle envelope lumen(GO:0031970) |
| 0.1 | 9.5 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 0.9 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 1.7 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 19.5 | GO:0045121 | membrane raft(GO:0045121) |
| 0.0 | 1.2 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 7.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 1.1 | GO:1990752 | microtubule end(GO:1990752) |
| 0.0 | 0.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 3.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.3 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 8.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 8.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 6.4 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 2.7 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 2.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 11.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 3.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 2.5 | GO:0030139 | endocytic vesicle(GO:0030139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.3 | 22.0 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 2.9 | 11.6 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 2.7 | 10.8 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 2.3 | 20.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 1.8 | 14.7 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 1.6 | 4.7 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 1.5 | 6.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 1.5 | 7.4 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 1.5 | 8.8 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 1.1 | 4.5 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 1.0 | 4.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.9 | 3.7 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.7 | 11.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.7 | 2.0 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.6 | 14.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.6 | 1.7 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.5 | 2.7 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.5 | 4.3 | GO:0038064 | collagen receptor activity(GO:0038064) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.4 | 3.9 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.4 | 3.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.4 | 1.6 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.3 | 1.0 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.3 | 11.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.3 | 0.9 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.3 | 5.8 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.3 | 2.4 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.3 | 3.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 1.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 3.7 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 5.5 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.2 | 3.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 0.6 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.2 | 0.7 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.2 | 2.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 3.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.6 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.6 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.6 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 7.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 3.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 5.0 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 8.2 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 0.3 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 11.9 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 1.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 15.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 4.6 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 2.7 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 2.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 1.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.6 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.9 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 1.5 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 0.7 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 1.0 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 2.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 2.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 2.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.0 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 20.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 2.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 1.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 4.1 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.9 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.3 | GO:0031701 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 2.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 3.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 5.0 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.7 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.5 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 11.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.4 | 20.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.3 | 14.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 5.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 5.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 0.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 4.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 4.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 3.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 3.2 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 13.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 4.0 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 2.1 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.6 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.4 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 10.8 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.4 | 14.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 11.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.3 | 6.8 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.2 | 7.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.2 | 4.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.2 | 4.4 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 4.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 3.9 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 6.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 3.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.0 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 1.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 4.0 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 0.7 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 3.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 6.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 0.9 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 1.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 2.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 3.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 5.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 1.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |