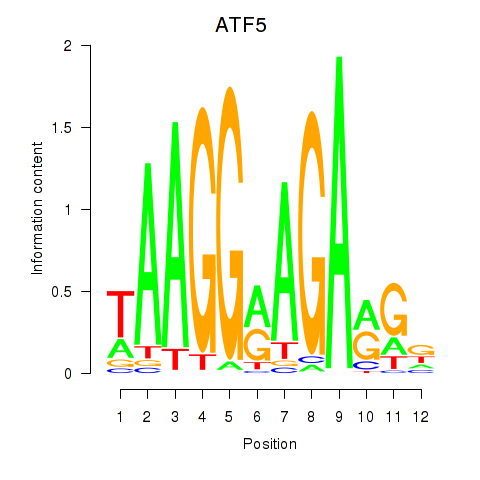
|
chrX_-_19965142
Show fit
|
23.22 |
ENST00000340625.3
|
BCLAF3
|
BCLAF1 and THRAP3 family member 3
|
|
chr11_+_92224801
Show fit
|
15.68 |
ENST00000525166.6
|
FAT3
|
FAT atypical cadherin 3
|
|
chr12_-_12338674
Show fit
|
11.78 |
ENST00000545735.1
|
MANSC1
|
MANSC domain containing 1
|
|
chr14_+_92323154
Show fit
|
11.56 |
ENST00000532405.6
ENST00000676001.1
ENST00000531433.5
|
SLC24A4
|
solute carrier family 24 member 4
|
|
chr21_-_26843063
Show fit
|
7.34 |
ENST00000678221.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1
|
|
chr21_-_26843012
Show fit
|
7.29 |
ENST00000517777.6
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1
|
|
chr19_+_41003946
Show fit
|
6.10 |
ENST00000593831.1
|
CYP2B6
|
cytochrome P450 family 2 subfamily B member 6
|
|
chr1_-_145095528
Show fit
|
6.09 |
ENST00000612199.4
ENST00000641863.1
|
SRGAP2B
|
SLIT-ROBO Rho GTPase activating protein 2B
|
|
chr12_+_53098846
Show fit
|
5.64 |
ENST00000650247.1
ENST00000549628.1
|
IGFBP6
|
insulin like growth factor binding protein 6
|
|
chr1_+_121184964
Show fit
|
5.33 |
ENST00000367123.8
|
SRGAP2C
|
SLIT-ROBO Rho GTPase activating protein 2C
|
|
chr8_+_22605018
Show fit
|
4.76 |
ENST00000389279.7
|
CCAR2
|
cell cycle and apoptosis regulator 2
|
|
chr8_+_22604632
Show fit
|
4.17 |
ENST00000308511.8
ENST00000523801.5
ENST00000521301.5
|
CCAR2
|
cell cycle and apoptosis regulator 2
|
|
chr5_+_83471764
Show fit
|
4.01 |
ENST00000512590.6
ENST00000513960.5
ENST00000513984.5
|
VCAN
|
versican
|
|
chr1_+_15756659
Show fit
|
3.98 |
ENST00000375771.5
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr2_+_188974364
Show fit
|
3.91 |
ENST00000304636.9
ENST00000317840.9
|
COL3A1
|
collagen type III alpha 1 chain
|
|
chr6_+_71886703
Show fit
|
3.84 |
ENST00000491071.6
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr22_-_35840577
Show fit
|
3.79 |
ENST00000405409.6
|
RBFOX2
|
RNA binding fox-1 homolog 2
|
|
chr15_-_58014097
Show fit
|
3.70 |
ENST00000559517.5
|
ALDH1A2
|
aldehyde dehydrogenase 1 family member A2
|
|
chr5_+_83471668
Show fit
|
3.67 |
ENST00000342785.8
ENST00000343200.9
|
VCAN
|
versican
|
|
chr5_+_83471925
Show fit
|
3.63 |
ENST00000502527.2
|
VCAN
|
versican
|
|
chr2_-_112784486
Show fit
|
3.38 |
ENST00000263339.4
|
IL1A
|
interleukin 1 alpha
|
|
chrX_+_104566193
Show fit
|
3.35 |
ENST00000372582.6
|
IL1RAPL2
|
interleukin 1 receptor accessory protein like 2
|
|
chr5_+_83471736
Show fit
|
3.31 |
ENST00000265077.8
|
VCAN
|
versican
|
|
chr12_+_53380141
Show fit
|
3.09 |
ENST00000551969.5
ENST00000327443.9
|
SP1
|
Sp1 transcription factor
|
|
chr9_-_23826231
Show fit
|
3.03 |
ENST00000397312.7
|
ELAVL2
|
ELAV like RNA binding protein 2
|
|
chr3_-_99850976
Show fit
|
2.99 |
ENST00000487087.5
|
FILIP1L
|
filamin A interacting protein 1 like
|
|
chr1_+_209686173
Show fit
|
2.86 |
ENST00000615289.4
ENST00000367028.6
ENST00000261465.5
|
HSD11B1
|
hydroxysteroid 11-beta dehydrogenase 1
|
|
chr19_+_15049469
Show fit
|
2.85 |
ENST00000427043.4
|
CASP14
|
caspase 14
|
|
chr6_+_71886900
Show fit
|
2.79 |
ENST00000517960.5
ENST00000518273.5
ENST00000522291.5
ENST00000521978.5
ENST00000520567.5
ENST00000264839.11
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr1_+_151766655
Show fit
|
2.78 |
ENST00000400999.7
|
OAZ3
|
ornithine decarboxylase antizyme 3
|
|
chr9_+_2622053
Show fit
|
2.78 |
ENST00000681306.1
ENST00000681618.1
|
VLDLR
|
very low density lipoprotein receptor
|
|
chr2_+_72916183
Show fit
|
2.56 |
ENST00000394111.6
|
EMX1
|
empty spiracles homeobox 1
|
|
chrX_-_130385656
Show fit
|
2.56 |
ENST00000276218.4
ENST00000682440.1
|
GPR119
|
G protein-coupled receptor 119
|
|
chr12_-_110502065
Show fit
|
2.54 |
ENST00000447578.6
ENST00000546588.1
ENST00000360579.11
ENST00000549578.6
ENST00000549970.5
|
VPS29
|
VPS29 retromer complex component
|
|
chr1_+_42463221
Show fit
|
2.50 |
ENST00000654683.1
ENST00000667205.1
ENST00000655164.1
ENST00000657597.1
ENST00000667947.1
ENST00000668663.1
ENST00000660083.1
ENST00000655845.1
ENST00000671281.1
ENST00000664805.1
ENST00000654604.1
ENST00000655447.1
ENST00000661864.1
ENST00000665176.1
ENST00000670982.1
ENST00000668036.1
|
CCDC30
|
coiled-coil domain containing 30
|
|
chr2_-_187554473
Show fit
|
2.46 |
ENST00000453013.5
ENST00000417013.5
|
TFPI
|
tissue factor pathway inhibitor
|
|
chr4_-_82798735
Show fit
|
2.43 |
ENST00000273908.4
ENST00000319540.9
|
SCD5
|
stearoyl-CoA desaturase 5
|
|
chr7_-_42152444
Show fit
|
2.43 |
ENST00000479210.1
|
GLI3
|
GLI family zinc finger 3
|
|
chr12_+_110502307
Show fit
|
2.42 |
ENST00000409778.7
ENST00000409300.6
|
RAD9B
|
RAD9 checkpoint clamp component B
|
|
chr1_-_183418364
Show fit
|
2.37 |
ENST00000287713.7
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr7_-_42152396
Show fit
|
2.34 |
ENST00000642432.1
ENST00000647255.1
ENST00000677288.1
|
GLI3
|
GLI family zinc finger 3
|
|
chr9_+_2621766
Show fit
|
2.30 |
ENST00000382100.8
|
VLDLR
|
very low density lipoprotein receptor
|
|
chr19_-_14979848
Show fit
|
2.24 |
ENST00000594383.2
|
SLC1A6
|
solute carrier family 1 member 6
|
|
chr18_-_812516
Show fit
|
2.20 |
ENST00000584307.5
|
YES1
|
YES proto-oncogene 1, Src family tyrosine kinase
|
|
chr6_+_101181254
Show fit
|
2.13 |
ENST00000682090.1
ENST00000421544.6
|
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2
|
|
chr10_-_102419934
Show fit
|
2.13 |
ENST00000406432.5
|
PSD
|
pleckstrin and Sec7 domain containing
|
|
chr1_-_16978276
Show fit
|
2.08 |
ENST00000375534.7
|
MFAP2
|
microfibril associated protein 2
|
|
chr22_+_19131271
Show fit
|
2.05 |
ENST00000399635.4
|
TSSK2
|
testis specific serine kinase 2
|
|
chr12_-_16608073
Show fit
|
2.04 |
ENST00000441439.6
|
LMO3
|
LIM domain only 3
|
|
chr17_+_35587478
Show fit
|
2.03 |
ENST00000618940.4
|
AP2B1
|
adaptor related protein complex 2 subunit beta 1
|
|
chr12_-_52573816
Show fit
|
2.02 |
ENST00000549343.5
ENST00000305620.3
|
KRT74
|
keratin 74
|
|
chr12_+_32106762
Show fit
|
2.01 |
ENST00000551848.1
ENST00000652176.1
|
BICD1
|
BICD cargo adaptor 1
|
|
chr6_+_43770707
Show fit
|
1.86 |
ENST00000324450.11
ENST00000417285.7
ENST00000413642.8
ENST00000372055.9
ENST00000482630.7
ENST00000425836.7
ENST00000372064.9
ENST00000372077.8
ENST00000519767.5
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr22_-_35961623
Show fit
|
1.84 |
ENST00000408983.2
|
RBFOX2
|
RNA binding fox-1 homolog 2
|
|
chr4_+_83456045
Show fit
|
1.81 |
ENST00000507349.5
ENST00000295491.9
ENST00000509970.5
ENST00000505719.1
|
MRPS18C
|
mitochondrial ribosomal protein S18C
|
|
chr7_+_128758947
Show fit
|
1.79 |
ENST00000493278.1
|
CALU
|
calumenin
|
|
chr6_-_62286161
Show fit
|
1.78 |
ENST00000281156.5
|
KHDRBS2
|
KH RNA binding domain containing, signal transduction associated 2
|
|
chr6_-_31546552
Show fit
|
1.78 |
ENST00000303892.10
ENST00000376151.4
|
ATP6V1G2
|
ATPase H+ transporting V1 subunit G2
|
|
chr10_-_102419693
Show fit
|
1.77 |
ENST00000611678.4
|
PSD
|
pleckstrin and Sec7 domain containing
|
|
chr12_-_16608183
Show fit
|
1.76 |
ENST00000354662.5
ENST00000538051.5
|
LMO3
|
LIM domain only 3
|
|
chr5_-_20575850
Show fit
|
1.73 |
ENST00000507958.5
|
CDH18
|
cadherin 18
|
|
chr2_-_25982471
Show fit
|
1.70 |
ENST00000264712.8
|
KIF3C
|
kinesin family member 3C
|
|
chr2_-_26478113
Show fit
|
1.66 |
ENST00000339598.8
|
OTOF
|
otoferlin
|
|
chr15_+_80441229
Show fit
|
1.66 |
ENST00000533983.5
ENST00000527771.5
ENST00000525103.1
|
ARNT2
|
aryl hydrocarbon receptor nuclear translocator 2
|
|
chr18_-_812230
Show fit
|
1.65 |
ENST00000314574.5
|
YES1
|
YES proto-oncogene 1, Src family tyrosine kinase
|
|
chr17_-_48614628
Show fit
|
1.64 |
ENST00000576562.1
|
HOXB8
|
homeobox B8
|
|
chr1_-_151346806
Show fit
|
1.63 |
ENST00000392746.7
|
RFX5
|
regulatory factor X5
|
|
chr15_+_74826603
Show fit
|
1.63 |
ENST00000395018.6
|
CPLX3
|
complexin 3
|
|
chr16_-_30012294
Show fit
|
1.58 |
ENST00000564979.5
ENST00000563378.5
|
DOC2A
|
double C2 domain alpha
|
|
chrX_+_155071414
Show fit
|
1.57 |
ENST00000340647.8
|
BRCC3
|
BRCA1/BRCA2-containing complex subunit 3
|
|
chr12_+_56521951
Show fit
|
1.55 |
ENST00000552247.6
|
RBMS2
|
RNA binding motif single stranded interacting protein 2
|
|
chrY_+_12904102
Show fit
|
1.51 |
ENST00000360160.8
ENST00000454054.5
|
DDX3Y
|
DEAD-box helicase 3 Y-linked
|
|
chr6_+_31897775
Show fit
|
1.50 |
ENST00000469372.5
ENST00000497706.5
|
C2
|
complement C2
|
|
chrX_-_129523436
Show fit
|
1.49 |
ENST00000371123.5
ENST00000371121.5
ENST00000371122.8
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr2_-_187554351
Show fit
|
1.49 |
ENST00000437725.5
ENST00000409676.5
ENST00000233156.9
ENST00000339091.8
ENST00000420747.1
|
TFPI
|
tissue factor pathway inhibitor
|
|
chr2_-_190062721
Show fit
|
1.48 |
ENST00000260950.5
|
MSTN
|
myostatin
|
|
chr4_-_121164314
Show fit
|
1.48 |
ENST00000057513.8
|
TNIP3
|
TNFAIP3 interacting protein 3
|
|
chr20_+_56412112
Show fit
|
1.47 |
ENST00000360314.7
|
CASS4
|
Cas scaffold protein family member 4
|
|
chr1_+_3690654
Show fit
|
1.46 |
ENST00000378285.5
ENST00000378280.5
ENST00000378288.8
|
TP73
|
tumor protein p73
|
|
chr22_-_35840218
Show fit
|
1.45 |
ENST00000414461.6
ENST00000416721.6
ENST00000449924.6
ENST00000262829.11
ENST00000397305.3
|
RBFOX2
|
RNA binding fox-1 homolog 2
|
|
chr11_-_129192291
Show fit
|
1.44 |
ENST00000682385.1
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr20_+_56412249
Show fit
|
1.43 |
ENST00000679887.1
ENST00000434344.2
|
CASS4
|
Cas scaffold protein family member 4
|
|
chr6_+_87155537
Show fit
|
1.40 |
ENST00000369577.8
ENST00000518845.1
ENST00000339907.8
ENST00000496806.2
|
ZNF292
|
zinc finger protein 292
|
|
chr17_-_35943662
Show fit
|
1.38 |
ENST00000618542.4
|
LYZL6
|
lysozyme like 6
|
|
chr12_+_56521990
Show fit
|
1.37 |
ENST00000550726.5
ENST00000542360.1
|
RBMS2
|
RNA binding motif single stranded interacting protein 2
|
|
chr1_+_35557435
Show fit
|
1.36 |
ENST00000373253.7
|
NCDN
|
neurochondrin
|
|
chr7_+_136869077
Show fit
|
1.36 |
ENST00000320658.9
ENST00000453373.5
ENST00000401861.1
|
CHRM2
|
cholinergic receptor muscarinic 2
|
|
chr11_+_63888515
Show fit
|
1.35 |
ENST00000509502.6
ENST00000512060.1
|
MARK2
|
microtubule affinity regulating kinase 2
|
|
chr14_+_70452161
Show fit
|
1.34 |
ENST00000603540.2
|
ADAM21
|
ADAM metallopeptidase domain 21
|
|
chr19_-_18255385
Show fit
|
1.33 |
ENST00000355502.7
ENST00000643046.1
|
ENSG00000285188.1
ENSG00000284797.1
|
novel protein
novel protein
|
|
chr2_-_113241683
Show fit
|
1.32 |
ENST00000468980.3
|
PAX8
|
paired box 8
|
|
chr17_-_35943707
Show fit
|
1.28 |
ENST00000615905.5
|
LYZL6
|
lysozyme like 6
|
|
chr11_+_118606428
Show fit
|
1.26 |
ENST00000361417.6
|
PHLDB1
|
pleckstrin homology like domain family B member 1
|
|
chr2_+_172860038
Show fit
|
1.23 |
ENST00000538974.5
ENST00000540783.5
|
RAPGEF4
|
Rap guanine nucleotide exchange factor 4
|
|
chr2_-_55050518
Show fit
|
1.22 |
ENST00000317610.11
ENST00000357732.8
|
RTN4
|
reticulon 4
|
|
chr2_-_100142575
Show fit
|
1.22 |
ENST00000317233.8
ENST00000672204.1
ENST00000416492.5
ENST00000672857.1
ENST00000672756.2
|
AFF3
|
AF4/FMR2 family member 3
|
|
chr16_-_58629816
Show fit
|
1.19 |
ENST00000564557.1
ENST00000317147.10
ENST00000569240.5
ENST00000441024.6
|
CNOT1
|
CCR4-NOT transcription complex subunit 1
|
|
chr20_+_56412393
Show fit
|
1.19 |
ENST00000679529.1
|
CASS4
|
Cas scaffold protein family member 4
|
|
chr11_-_84720876
Show fit
|
1.15 |
ENST00000648622.1
|
DLG2
|
discs large MAGUK scaffold protein 2
|
|
chr19_+_4229502
Show fit
|
1.14 |
ENST00000221847.6
|
EBI3
|
Epstein-Barr virus induced 3
|
|
chrX_+_154144242
Show fit
|
1.14 |
ENST00000369951.9
|
OPN1LW
|
opsin 1, long wave sensitive
|
|
chr18_-_55422492
Show fit
|
1.12 |
ENST00000561992.5
ENST00000630712.2
|
TCF4
|
transcription factor 4
|
|
chr8_-_13276491
Show fit
|
1.10 |
ENST00000512044.6
|
DLC1
|
DLC1 Rho GTPase activating protein
|
|
chr4_-_174829212
Show fit
|
1.10 |
ENST00000340217.5
ENST00000274093.8
|
GLRA3
|
glycine receptor alpha 3
|
|
chr14_+_79279403
Show fit
|
1.07 |
ENST00000281127.11
|
NRXN3
|
neurexin 3
|
|
chr1_-_11803383
Show fit
|
1.07 |
ENST00000641407.1
ENST00000376583.7
ENST00000423400.7
|
MTHFR
|
methylenetetrahydrofolate reductase
|
|
chr2_-_113241779
Show fit
|
1.06 |
ENST00000497038.6
|
PAX8
|
paired box 8
|
|
chr14_+_79279681
Show fit
|
1.03 |
ENST00000679122.1
|
NRXN3
|
neurexin 3
|
|
chr11_-_129192198
Show fit
|
1.03 |
ENST00000310343.13
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr9_+_19230680
Show fit
|
1.02 |
ENST00000434457.7
|
DENND4C
|
DENN domain containing 4C
|
|
chr16_-_72172135
Show fit
|
1.01 |
ENST00000537465.5
ENST00000237353.15
|
PMFBP1
|
polyamine modulated factor 1 binding protein 1
|
|
chr12_-_24949026
Show fit
|
1.00 |
ENST00000539780.5
ENST00000546285.1
ENST00000342945.9
ENST00000261192.12
|
BCAT1
|
branched chain amino acid transaminase 1
|
|
chr4_+_165873231
Show fit
|
0.99 |
ENST00000061240.7
|
TLL1
|
tolloid like 1
|
|
chr12_-_56449377
Show fit
|
0.99 |
ENST00000229201.4
ENST00000553532.6
|
TIMELESS
|
timeless circadian regulator
|
|
chr1_+_35557768
Show fit
|
0.97 |
ENST00000356090.8
ENST00000373243.7
|
NCDN
|
neurochondrin
|
|
chr5_+_132673983
Show fit
|
0.97 |
ENST00000622422.1
ENST00000231449.7
ENST00000350025.2
|
IL4
|
interleukin 4
|
|
chr12_+_32107151
Show fit
|
0.93 |
ENST00000548411.5
|
BICD1
|
BICD cargo adaptor 1
|
|
chr8_-_21788159
Show fit
|
0.92 |
ENST00000517892.5
ENST00000518077.5
|
GFRA2
|
GDNF family receptor alpha 2
|
|
chrX_+_154182596
Show fit
|
0.92 |
ENST00000595290.6
|
OPN1MW
|
opsin 1, medium wave sensitive
|
|
chr19_-_54360949
Show fit
|
0.91 |
ENST00000622064.1
|
LAIR1
|
leukocyte associated immunoglobulin like receptor 1
|
|
chr22_-_36819479
Show fit
|
0.91 |
ENST00000216200.9
|
PVALB
|
parvalbumin
|
|
chr1_-_156490599
Show fit
|
0.90 |
ENST00000360595.7
|
MEF2D
|
myocyte enhancer factor 2D
|
|
chr3_-_187736493
Show fit
|
0.90 |
ENST00000232014.8
|
BCL6
|
BCL6 transcription repressor
|
|
chrX_+_155071473
Show fit
|
0.89 |
ENST00000369459.6
ENST00000369462.5
ENST00000330045.12
ENST00000411985.5
ENST00000399042.4
ENST00000620502.4
ENST00000630295.1
|
BRCC3
|
BRCA1/BRCA2-containing complex subunit 3
|
|
chr18_-_34224871
Show fit
|
0.88 |
ENST00000261592.10
|
NOL4
|
nucleolar protein 4
|
|
chr4_+_143336762
Show fit
|
0.88 |
ENST00000262995.8
|
GAB1
|
GRB2 associated binding protein 1
|
|
chr8_-_21788853
Show fit
|
0.87 |
ENST00000524240.6
|
GFRA2
|
GDNF family receptor alpha 2
|
|
chr17_+_7630094
Show fit
|
0.87 |
ENST00000441599.6
ENST00000380450.9
ENST00000416273.7
ENST00000575903.5
ENST00000571153.5
ENST00000575618.5
ENST00000576152.1
ENST00000576830.5
|
SHBG
|
sex hormone binding globulin
|
|
chr17_-_7205116
Show fit
|
0.87 |
ENST00000649520.1
ENST00000649186.1
|
DLG4
|
discs large MAGUK scaffold protein 4
|
|
chr4_+_76251694
Show fit
|
0.86 |
ENST00000510328.5
ENST00000424749.7
ENST00000502320.2
ENST00000515604.5
|
FAM47E
FAM47E-STBD1
|
family with sequence similarity 47 member E
FAM47E-STBD1 readthrough
|
|
chr12_+_53380639
Show fit
|
0.86 |
ENST00000426431.2
|
SP1
|
Sp1 transcription factor
|
|
chr3_+_19947316
Show fit
|
0.85 |
ENST00000422242.1
|
RAB5A
|
RAB5A, member RAS oncogene family
|
|
chr9_+_19230435
Show fit
|
0.81 |
ENST00000602925.5
|
DENND4C
|
DENN domain containing 4C
|
|
chr5_+_111223905
Show fit
|
0.81 |
ENST00000512453.5
|
CAMK4
|
calcium/calmodulin dependent protein kinase IV
|
|
chr17_+_4771878
Show fit
|
0.81 |
ENST00000270560.4
|
TM4SF5
|
transmembrane 4 L six family member 5
|
|
chr12_-_71663827
Show fit
|
0.80 |
ENST00000378743.9
ENST00000552037.1
|
ZFC3H1
|
zinc finger C3H1-type containing
|
|
chrX_+_123184260
Show fit
|
0.79 |
ENST00000620443.2
ENST00000622768.5
ENST00000611689.4
|
GRIA3
|
glutamate ionotropic receptor AMPA type subunit 3
|
|
chr2_+_27051637
Show fit
|
0.79 |
ENST00000451003.5
ENST00000360131.5
ENST00000323064.12
|
AGBL5
|
ATP/GTP binding protein like 5
|
|
chr19_+_42269219
Show fit
|
0.79 |
ENST00000681038.1
|
CIC
|
capicua transcriptional repressor
|
|
chrX_+_12137409
Show fit
|
0.78 |
ENST00000672010.1
|
FRMPD4
|
FERM and PDZ domain containing 4
|
|
chr10_+_101354028
Show fit
|
0.78 |
ENST00000393441.8
|
BTRC
|
beta-transducin repeat containing E3 ubiquitin protein ligase
|
|
chr1_+_206203541
Show fit
|
0.78 |
ENST00000573034.8
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2
|
|
chr2_-_208129824
Show fit
|
0.76 |
ENST00000282141.4
|
CRYGC
|
crystallin gamma C
|
|
chr13_-_27969295
Show fit
|
0.75 |
ENST00000381020.8
|
CDX2
|
caudal type homeobox 2
|
|
chr18_-_55422306
Show fit
|
0.75 |
ENST00000566777.5
ENST00000626584.2
|
TCF4
|
transcription factor 4
|
|
chr6_+_29396555
Show fit
|
0.72 |
ENST00000623183.1
|
OR12D2
|
olfactory receptor family 12 subfamily D member 2
|
|
chrX_+_154219756
Show fit
|
0.72 |
ENST00000369929.8
|
OPN1MW2
|
opsin 1, medium wave sensitive 2
|
|
chr19_+_48465837
Show fit
|
0.71 |
ENST00000595676.1
|
ENSG00000268465.1
|
novel protein
|
|
chr10_+_101354083
Show fit
|
0.65 |
ENST00000408038.6
ENST00000370187.8
|
BTRC
|
beta-transducin repeat containing E3 ubiquitin protein ligase
|
|
chr12_+_56521798
Show fit
|
0.65 |
ENST00000262031.10
|
RBMS2
|
RNA binding motif single stranded interacting protein 2
|
|
chr14_+_79280263
Show fit
|
0.63 |
ENST00000555387.1
|
NRXN3
|
neurexin 3
|
|
chr9_+_79573162
Show fit
|
0.63 |
ENST00000425506.5
|
TLE4
|
TLE family member 4, transcriptional corepressor
|
|
chr4_-_67754335
Show fit
|
0.62 |
ENST00000420975.2
ENST00000226413.5
|
GNRHR
|
gonadotropin releasing hormone receptor
|
|
chr17_+_42853232
Show fit
|
0.59 |
ENST00000617500.4
|
AOC3
|
amine oxidase copper containing 3
|
|
chr3_-_183427977
Show fit
|
0.58 |
ENST00000473233.5
|
MCF2L2
|
MCF.2 cell line derived transforming sequence-like 2
|
|
chr18_-_3845292
Show fit
|
0.57 |
ENST00000400145.6
|
DLGAP1
|
DLG associated protein 1
|
|
chr15_+_63189554
Show fit
|
0.56 |
ENST00000559006.1
ENST00000321437.9
|
RAB8B
|
RAB8B, member RAS oncogene family
|
|
chr14_+_79279906
Show fit
|
0.54 |
ENST00000428277.6
|
NRXN3
|
neurexin 3
|
|
chr8_-_60281388
Show fit
|
0.54 |
ENST00000317995.5
|
CA8
|
carbonic anhydrase 8
|
|
chr4_+_70242583
Show fit
|
0.53 |
ENST00000304954.3
|
CSN3
|
casein kappa
|
|
chr11_+_123525822
Show fit
|
0.50 |
ENST00000322282.11
ENST00000529750.5
|
GRAMD1B
|
GRAM domain containing 1B
|
|
chr19_-_45370384
Show fit
|
0.47 |
ENST00000485403.6
ENST00000586856.1
ENST00000586131.6
ENST00000391945.10
ENST00000684407.1
ENST00000391944.8
|
ERCC2
|
ERCC excision repair 2, TFIIH core complex helicase subunit
|
|
chr1_+_158353965
Show fit
|
0.47 |
ENST00000368154.5
ENST00000368155.7
ENST00000368156.5
ENST00000368157.5
ENST00000368160.7
ENST00000368161.7
|
CD1E
|
CD1e molecule
|
|
chr8_+_78516329
Show fit
|
0.47 |
ENST00000396418.7
ENST00000352966.9
|
PKIA
|
cAMP-dependent protein kinase inhibitor alpha
|
|
chr5_-_149063021
Show fit
|
0.43 |
ENST00000515425.6
|
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2
|
|
chr8_-_109680812
Show fit
|
0.42 |
ENST00000528716.5
ENST00000527600.5
ENST00000531230.5
ENST00000532189.5
ENST00000534184.5
ENST00000408889.7
ENST00000533171.5
|
SYBU
|
syntabulin
|
|
chrX_-_13817346
Show fit
|
0.42 |
ENST00000356942.9
|
GPM6B
|
glycoprotein M6B
|
|
chr17_-_43125450
Show fit
|
0.42 |
ENST00000494123.5
ENST00000468300.5
ENST00000471181.7
ENST00000652672.1
|
BRCA1
|
BRCA1 DNA repair associated
|
|
chr8_-_42207557
Show fit
|
0.41 |
ENST00000220809.9
ENST00000429089.6
ENST00000519510.5
ENST00000429710.6
ENST00000524009.5
|
PLAT
|
plasminogen activator, tissue type
|
|
chr5_+_137867852
Show fit
|
0.40 |
ENST00000421631.6
ENST00000239926.9
|
MYOT
|
myotilin
|
|
chr12_-_114683590
Show fit
|
0.39 |
ENST00000257566.7
|
TBX3
|
T-box transcription factor 3
|
|
chr3_-_101513175
Show fit
|
0.36 |
ENST00000394091.5
ENST00000394094.6
ENST00000394095.7
ENST00000348610.3
ENST00000314261.11
|
SENP7
|
SUMO specific peptidase 7
|
|
chr6_-_32176051
Show fit
|
0.34 |
ENST00000375107.8
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1
|
|
chr6_-_160664270
Show fit
|
0.33 |
ENST00000316300.10
|
LPA
|
lipoprotein(a)
|
|
chr1_-_103108468
Show fit
|
0.32 |
ENST00000512756.5
ENST00000461720.6
ENST00000358392.6
ENST00000353414.8
|
COL11A1
|
collagen type XI alpha 1 chain
|
|
chr21_-_43058941
Show fit
|
0.31 |
ENST00000451248.5
ENST00000458223.5
|
CBS
|
cystathionine beta-synthase
|
|
chr14_+_79280056
Show fit
|
0.30 |
ENST00000676811.1
|
NRXN3
|
neurexin 3
|
|
chrX_-_13817027
Show fit
|
0.29 |
ENST00000493677.5
ENST00000355135.6
ENST00000316715.9
|
GPM6B
|
glycoprotein M6B
|
|
chr12_-_23951020
Show fit
|
0.28 |
ENST00000441133.2
ENST00000545921.5
|
SOX5
|
SRY-box transcription factor 5
|
|
chr1_+_202122910
Show fit
|
0.26 |
ENST00000682545.1
ENST00000367282.6
ENST00000682887.1
|
GPR37L1
|
G protein-coupled receptor 37 like 1
|
|
chr12_+_120446418
Show fit
|
0.25 |
ENST00000551765.6
ENST00000229384.5
|
GATC
|
glutamyl-tRNA amidotransferase subunit C
|
|
chr8_-_94208548
Show fit
|
0.24 |
ENST00000027335.8
ENST00000441892.6
ENST00000521491.1
|
CDH17
|
cadherin 17
|
|
chr1_-_46132650
Show fit
|
0.23 |
ENST00000372006.5
ENST00000425892.2
ENST00000420542.5
|
PIK3R3
|
phosphoinositide-3-kinase regulatory subunit 3
|
|
chr11_+_63681444
Show fit
|
0.23 |
ENST00000341307.6
ENST00000356000.7
ENST00000542238.5
|
RTN3
|
reticulon 3
|
|
chr11_+_63681483
Show fit
|
0.22 |
ENST00000339997.8
ENST00000540798.5
ENST00000545432.5
ENST00000543552.5
ENST00000377819.10
ENST00000537981.5
|
RTN3
|
reticulon 3
|
|
chr15_-_75579248
Show fit
|
0.22 |
ENST00000306726.6
ENST00000618819.5
|
PTPN9
|
protein tyrosine phosphatase non-receptor type 9
|
|
chr17_-_43125353
Show fit
|
0.21 |
ENST00000476777.5
ENST00000491747.6
ENST00000478531.5
ENST00000357654.9
ENST00000477152.5
ENST00000618469.1
ENST00000352993.7
ENST00000493795.5
ENST00000493919.5
|
BRCA1
|
BRCA1 DNA repair associated
|
|
chr10_-_73655984
Show fit
|
0.20 |
ENST00000394810.3
|
SYNPO2L
|
synaptopodin 2 like
|
|
chr12_-_114684151
Show fit
|
0.20 |
ENST00000349155.7
|
TBX3
|
T-box transcription factor 3
|
|
chr16_-_72093598
Show fit
|
0.15 |
ENST00000268483.8
|
TXNL4B
|
thioredoxin like 4B
|
|
chr2_-_87021844
Show fit
|
0.15 |
ENST00000355705.4
ENST00000409310.6
|
PLGLB1
|
plasminogen like B1
|
|
chr12_-_56636318
Show fit
|
0.13 |
ENST00000549506.5
ENST00000379441.7
ENST00000551812.5
|
BAZ2A
|
bromodomain adjacent to zinc finger domain 2A
|
|
chr1_-_158330957
Show fit
|
0.11 |
ENST00000451207.5
|
CD1B
|
CD1b molecule
|
|
chr15_-_23647848
Show fit
|
0.10 |
ENST00000650528.1
|
MAGEL2
|
MAGE family member L2
|
|
chr5_+_155013755
Show fit
|
0.09 |
ENST00000435029.6
|
KIF4B
|
kinesin family member 4B
|
|
chr1_+_202122881
Show fit
|
0.07 |
ENST00000683302.1
ENST00000683557.1
|
GPR37L1
|
G protein-coupled receptor 37 like 1
|
|
chr1_-_54406385
Show fit
|
0.06 |
ENST00000610401.5
|
SSBP3
|
single stranded DNA binding protein 3
|
|
chr5_+_137867868
Show fit
|
0.03 |
ENST00000515645.1
|
MYOT
|
myotilin
|
|
chr8_+_99013247
Show fit
|
0.01 |
ENST00000441350.2
ENST00000357162.7
ENST00000358544.7
|
VPS13B
|
vacuolar protein sorting 13 homolog B
|