Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for BACH1_NFE2_NFE2L2
Z-value: 10.58
Transcription factors associated with BACH1_NFE2_NFE2L2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
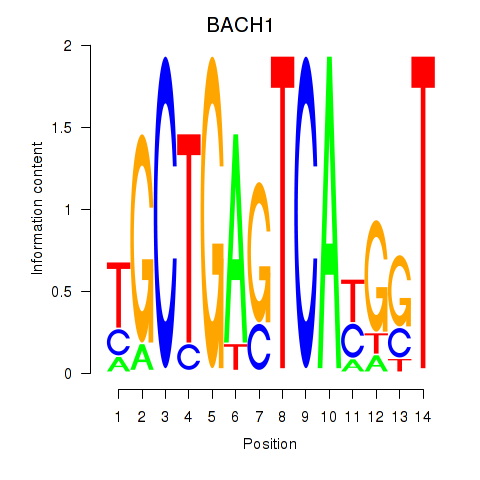
BACH1
|
ENSG00000156273.16 | BACH1 |
|
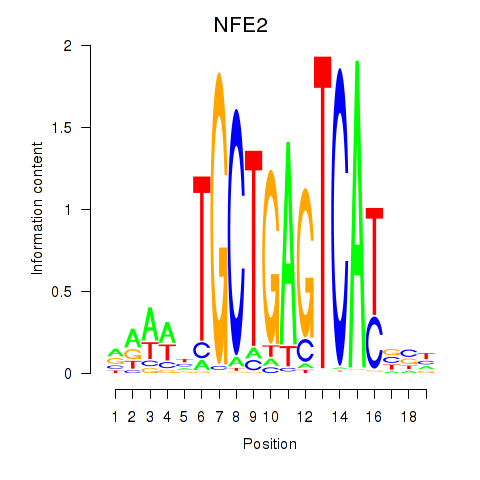
NFE2
|
ENSG00000123405.14 | NFE2 |
|
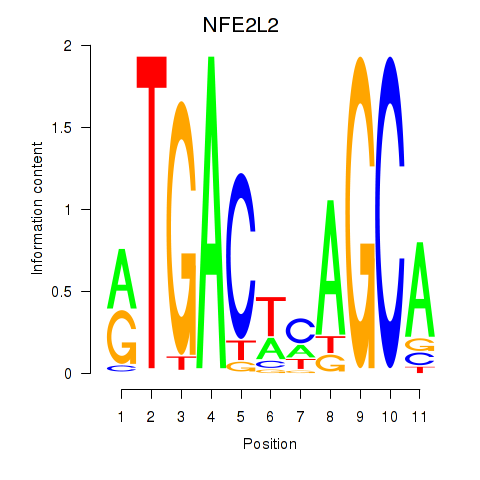
NFE2L2
|
ENSG00000116044.16 | NFE2L2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFE2L2 | hg38_v1_chr2_-_177392673_177392704, hg38_v1_chr2_-_177263800_177263870, hg38_v1_chr2_-_177263522_177263572, hg38_v1_chr2_-_177264686_177264823 | 0.29 | 1.8e-05 | Click! |
| BACH1 | hg38_v1_chr21_+_29298890_29298932, hg38_v1_chr21_+_29299368_29299441 | -0.20 | 3.5e-03 | Click! |
| NFE2 | hg38_v1_chr12_-_54297884_54297941 | -0.02 | 7.2e-01 | Click! |
Activity profile of BACH1_NFE2_NFE2L2 motif
Sorted Z-values of BACH1_NFE2_NFE2L2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of BACH1_NFE2_NFE2L2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 24.1 | 72.2 | GO:0014810 | positive regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014810) |
| 21.0 | 84.0 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 17.3 | 52.0 | GO:1904387 | response to human chorionic gonadotropin(GO:0044752) cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 15.3 | 46.0 | GO:1901388 | regulation of transforming growth factor beta activation(GO:1901388) negative regulation of transforming growth factor beta activation(GO:1901389) |
| 14.0 | 56.0 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 12.8 | 38.3 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 9.5 | 28.5 | GO:0035732 | nitric oxide storage(GO:0035732) |
| 7.6 | 30.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 7.5 | 52.4 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 7.4 | 773.0 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 7.4 | 73.8 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 6.8 | 20.4 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 6.4 | 51.0 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 6.2 | 18.6 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 6.0 | 24.2 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 5.5 | 33.1 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 5.3 | 96.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 5.3 | 16.0 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) regulation of chromatin silencing at telomere(GO:0031938) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 5.0 | 25.0 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 4.4 | 17.8 | GO:1904020 | negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of G-protein coupled receptor internalization(GO:1904020) negative regulation of G-protein coupled receptor internalization(GO:1904021) |
| 3.9 | 11.7 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) |
| 3.8 | 11.5 | GO:0090176 | microtubule cytoskeleton organization involved in establishment of planar polarity(GO:0090176) |
| 3.7 | 11.2 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 3.6 | 32.7 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 3.6 | 122.6 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 3.4 | 20.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 3.4 | 34.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 3.0 | 21.2 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 3.0 | 41.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 2.9 | 25.8 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 2.6 | 13.1 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 2.6 | 12.9 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 2.5 | 14.8 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 2.5 | 32.0 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 2.4 | 31.6 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 2.4 | 21.4 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 2.2 | 24.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 2.2 | 44.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 2.2 | 13.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 2.2 | 28.1 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 1.9 | 7.5 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 1.8 | 29.8 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 1.6 | 4.9 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 1.6 | 4.9 | GO:1903004 | flavin adenine dinucleotide metabolic process(GO:0072387) regulation of protein K63-linked deubiquitination(GO:1903004) positive regulation of protein K63-linked deubiquitination(GO:1903006) |
| 1.4 | 15.7 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 1.4 | 9.9 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 1.4 | 23.6 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 1.4 | 6.8 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 1.4 | 5.4 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 1.3 | 8.0 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 1.3 | 7.6 | GO:0060356 | leucine import(GO:0060356) |
| 1.3 | 7.5 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 1.2 | 9.9 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 1.2 | 58.4 | GO:0043486 | histone exchange(GO:0043486) |
| 1.2 | 13.0 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 1.2 | 29.0 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 1.1 | 3.4 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 1.1 | 13.4 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 1.1 | 4.4 | GO:0002384 | hepatic immune response(GO:0002384) response to prolactin(GO:1990637) |
| 1.1 | 4.4 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 1.1 | 5.4 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 1.1 | 14.0 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 1.0 | 10.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 1.0 | 9.0 | GO:1902661 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) positive regulation of glucose mediated signaling pathway(GO:1902661) |
| 1.0 | 10.0 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 1.0 | 2.0 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 1.0 | 9.7 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 1.0 | 12.6 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 1.0 | 5.7 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.9 | 4.6 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.9 | 32.4 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.9 | 12.4 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.9 | 20.8 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.8 | 36.5 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.8 | 14.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.8 | 2.5 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.8 | 3.2 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.8 | 16.1 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.7 | 10.9 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.7 | 4.3 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.7 | 4.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.7 | 5.9 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) cellular response to interleukin-2(GO:0071352) |
| 0.6 | 8.4 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.6 | 7.6 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.6 | 36.0 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.6 | 9.5 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.6 | 5.3 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.5 | 39.3 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.5 | 27.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.5 | 2.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.5 | 2.1 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.5 | 3.2 | GO:0030421 | defecation(GO:0030421) |
| 0.4 | 2.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.4 | 13.7 | GO:0072574 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.4 | 1.6 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.4 | 42.3 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.4 | 13.6 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.3 | 4.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.3 | 9.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.3 | 1.6 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.3 | 15.8 | GO:0006294 | nucleotide-excision repair, preincision complex assembly(GO:0006294) |
| 0.3 | 11.6 | GO:0071173 | mitotic spindle assembly checkpoint(GO:0007094) spindle assembly checkpoint(GO:0071173) |
| 0.3 | 4.9 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.3 | 4.6 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.3 | 2.6 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.3 | 29.4 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.2 | 2.7 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.2 | 6.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.2 | 0.7 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.2 | 13.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.2 | 3.2 | GO:0000478 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) |
| 0.2 | 4.5 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.2 | 11.5 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.2 | 8.0 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.2 | 0.7 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 1.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 8.8 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 | 2.0 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 1.1 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 5.7 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.1 | 3.1 | GO:0033599 | regulation of mammary gland epithelial cell proliferation(GO:0033599) |
| 0.1 | 4.4 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 1.8 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.1 | 7.8 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 4.2 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.1 | 2.3 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.1 | 5.2 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 | 33.9 | GO:0007411 | axon guidance(GO:0007411) |
| 0.1 | 13.3 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.1 | 1.8 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 2.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 5.6 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 15.7 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.1 | 1.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 1.5 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.1 | 11.3 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.1 | 1.4 | GO:0002251 | organ or tissue specific immune response(GO:0002251) |
| 0.0 | 8.5 | GO:0060348 | bone development(GO:0060348) |
| 0.0 | 1.3 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 1.4 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 3.1 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 1.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 6.1 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.0 | 1.4 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.1 | GO:0045425 | regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.1 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) |
| 0.0 | 1.7 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 4.8 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 2.5 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 1.3 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 3.4 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 33.6 | 403.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 18.7 | 56.0 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 17.3 | 52.0 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 15.3 | 46.0 | GO:1990917 | sperm head plasma membrane(GO:1990913) ooplasm(GO:1990917) |
| 10.0 | 110.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 9.0 | 81.4 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 7.2 | 29.0 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 7.1 | 28.5 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 5.4 | 21.4 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 5.3 | 16.0 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 5.3 | 42.4 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 4.5 | 31.6 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 3.7 | 71.0 | GO:0000812 | Swr1 complex(GO:0000812) |
| 3.2 | 16.1 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 2.8 | 36.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 2.7 | 24.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 2.3 | 9.2 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 2.2 | 15.7 | GO:0044754 | autolysosome(GO:0044754) |
| 2.2 | 36.9 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 2.1 | 111.4 | GO:0000502 | proteasome complex(GO:0000502) |
| 2.1 | 20.8 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 2.0 | 42.4 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 2.0 | 41.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 1.9 | 5.7 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 1.8 | 33.1 | GO:0031045 | dense core granule(GO:0031045) |
| 1.8 | 32.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 1.6 | 144.0 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 1.5 | 24.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 1.3 | 12.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 1.2 | 11.5 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 1.1 | 14.0 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.9 | 14.4 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.9 | 4.4 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.8 | 2.5 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.8 | 29.8 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.8 | 8.4 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.7 | 132.9 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.6 | 86.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.5 | 34.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.5 | 4.3 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) ERCC4-ERCC1 complex(GO:0070522) |
| 0.5 | 6.8 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.4 | 2.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.4 | 21.5 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.4 | 13.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.4 | 9.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.4 | 8.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.3 | 7.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.3 | 13.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.3 | 10.7 | GO:0071564 | SWI/SNF complex(GO:0016514) npBAF complex(GO:0071564) |
| 0.3 | 54.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.3 | 25.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.3 | 50.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.3 | 13.7 | GO:0005776 | autophagosome(GO:0005776) |
| 0.3 | 5.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.3 | 4.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 13.6 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 11.3 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.2 | 2.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 13.9 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.2 | 3.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.2 | 9.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 61.1 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.2 | 7.5 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 2.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 2.0 | GO:0032059 | bleb(GO:0032059) |
| 0.1 | 3.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 11.1 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 4.1 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 5.4 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 28.5 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 4.8 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.4 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 19.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 17.9 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 34.0 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 18.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 5.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 120.1 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 3.3 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 10.4 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 2.0 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 21.0 | 84.0 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 17.3 | 52.0 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 15.8 | 47.5 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 15.7 | 125.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 15.3 | 46.0 | GO:0002135 | CTP binding(GO:0002135) |
| 15.0 | 421.0 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 14.7 | 88.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 14.4 | 72.2 | GO:0045174 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 14.0 | 56.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 11.6 | 115.8 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 9.5 | 28.5 | GO:0035731 | S-nitrosoglutathione binding(GO:0035730) dinitrosyl-iron complex binding(GO:0035731) |
| 9.0 | 81.4 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 8.7 | 96.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 8.4 | 75.6 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 7.2 | 29.0 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 6.8 | 20.4 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 6.2 | 18.6 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 6.1 | 30.3 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) oxaloacetate decarboxylase activity(GO:0008948) |
| 6.0 | 29.8 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 4.3 | 21.4 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 4.1 | 28.5 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 4.0 | 16.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 4.0 | 24.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 3.9 | 11.7 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) |
| 3.7 | 11.2 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 3.2 | 31.9 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 2.9 | 20.6 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 2.8 | 36.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 2.5 | 9.8 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 2.2 | 13.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 2.0 | 8.0 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 1.8 | 14.0 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 1.7 | 32.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 1.7 | 10.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 1.7 | 10.0 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 1.6 | 24.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 1.6 | 8.0 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 1.5 | 9.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 1.5 | 5.9 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 1.5 | 45.0 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 1.4 | 17.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 1.4 | 5.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 1.3 | 74.5 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 1.1 | 16.0 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 1.1 | 4.2 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 1.0 | 27.6 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.8 | 15.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.7 | 12.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.7 | 11.5 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.7 | 9.9 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.7 | 5.9 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.6 | 9.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.6 | 14.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.6 | 7.6 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.6 | 25.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.6 | 12.1 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.6 | 5.7 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.5 | 2.7 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.5 | 1.6 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.5 | 4.3 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.5 | 9.7 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.5 | 1.4 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.4 | 4.4 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.4 | 37.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.4 | 3.4 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.4 | 1.6 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.4 | 9.2 | GO:0001163 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.4 | 7.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.4 | 7.8 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.4 | 5.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.3 | 9.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.3 | 82.8 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.3 | 6.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.3 | 1.8 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.3 | 13.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.3 | 2.5 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.3 | 16.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 7.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 4.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.2 | 7.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 5.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 4.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.2 | 2.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.2 | 2.1 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.2 | 68.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.2 | 4.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 2.6 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.2 | 7.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 11.0 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 8.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 2.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 10.8 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.1 | 4.6 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 5.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 1.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 5.4 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 3.1 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 2.1 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.1 | 6.7 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 5.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 3.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 10.9 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 5.1 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 1.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 2.0 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 10.6 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 5.6 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 62.8 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 1.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 1.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.0 | GO:0005135 | interleukin-3 receptor binding(GO:0005135) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 116.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 1.6 | 114.5 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 1.4 | 63.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 1.3 | 57.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 1.2 | 101.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 1.2 | 45.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 1.1 | 12.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 1.0 | 13.7 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.9 | 108.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.6 | 35.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.6 | 45.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.6 | 10.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.6 | 40.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.5 | 47.6 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.5 | 15.7 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.5 | 26.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.4 | 9.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.4 | 8.4 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.4 | 9.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.4 | 6.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.4 | 29.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.3 | 7.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.3 | 14.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.3 | 11.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.3 | 28.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.3 | 26.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.3 | 5.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.3 | 6.7 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.2 | 17.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.2 | 9.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 3.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 5.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 4.1 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 8.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 7.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.6 | 773.0 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 5.5 | 98.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 4.4 | 148.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 4.1 | 36.5 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 3.9 | 116.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 3.5 | 112.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 3.1 | 90.0 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 1.8 | 32.7 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 1.7 | 95.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 1.7 | 18.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 1.3 | 36.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 1.2 | 47.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 1.1 | 17.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 1.1 | 16.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 1.0 | 14.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.8 | 9.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.8 | 21.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.7 | 27.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.7 | 18.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.7 | 9.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.7 | 11.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.6 | 33.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.6 | 16.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.6 | 6.8 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.5 | 15.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.5 | 6.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.5 | 25.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.4 | 9.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.4 | 50.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.4 | 7.8 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.4 | 13.7 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.3 | 27.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.3 | 8.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 5.3 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.3 | 5.7 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.3 | 9.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 19.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.2 | 46.1 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.2 | 36.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.2 | 4.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 6.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 2.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.2 | 3.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 18.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 4.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 17.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 2.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.5 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 12.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 0.7 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.1 | 1.8 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 1.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.7 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |