Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
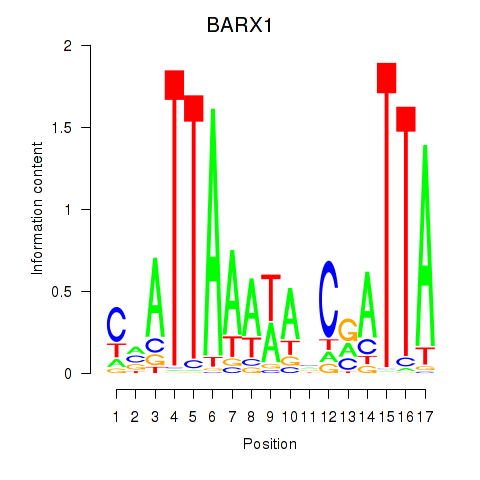
Results for BARX1
Z-value: 1.10
Transcription factors associated with BARX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BARX1
|
ENSG00000131668.14 | BARX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BARX1 | hg38_v1_chr9_-_93955347_93955362 | 0.11 | 1.2e-01 | Click! |
Activity profile of BARX1 motif
Sorted Z-values of BARX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of BARX1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.5 | 43.6 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 3.8 | 11.5 | GO:2000521 | negative regulation of immunological synapse formation(GO:2000521) |
| 3.4 | 10.1 | GO:0014873 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 2.1 | 6.3 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 1.8 | 5.5 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 1.7 | 6.7 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 1.6 | 6.5 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 1.5 | 7.5 | GO:1902019 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 1.5 | 16.1 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 1.5 | 7.3 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 1.4 | 9.8 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 1.4 | 5.4 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 1.3 | 4.0 | GO:0036292 | DNA rewinding(GO:0036292) |
| 1.2 | 3.7 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 1.2 | 7.0 | GO:1904304 | regulation of gastro-intestinal system smooth muscle contraction(GO:1904304) positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 1.1 | 10.1 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 1.1 | 13.2 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.1 | 14.0 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 1.0 | 4.1 | GO:0009137 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.9 | 4.3 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.8 | 1.7 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.8 | 4.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.8 | 2.4 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.7 | 5.2 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.7 | 9.6 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.7 | 4.4 | GO:0046086 | AMP catabolic process(GO:0006196) adenosine biosynthetic process(GO:0046086) |
| 0.7 | 2.2 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.7 | 9.4 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.7 | 5.7 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.7 | 7.0 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.7 | 13.9 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.7 | 4.6 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.6 | 1.9 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.6 | 4.5 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.6 | 3.1 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.6 | 3.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.6 | 2.5 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 0.6 | 3.6 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.6 | 5.7 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.6 | 4.5 | GO:0015705 | iodide transport(GO:0015705) |
| 0.6 | 10.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.6 | 7.7 | GO:0051964 | netrin-activated signaling pathway(GO:0038007) negative regulation of synapse assembly(GO:0051964) |
| 0.5 | 2.2 | GO:0036114 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.5 | 3.2 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.5 | 11.7 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.5 | 4.7 | GO:0060318 | definitive erythrocyte differentiation(GO:0060318) |
| 0.5 | 9.4 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.5 | 1.6 | GO:0019605 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 0.5 | 14.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.5 | 2.9 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.5 | 2.3 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) |
| 0.4 | 5.7 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.4 | 76.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.4 | 5.5 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.4 | 2.5 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.4 | 7.4 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.4 | 3.2 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.4 | 1.6 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.4 | 1.2 | GO:0070837 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) dehydroascorbic acid transport(GO:0070837) |
| 0.4 | 2.7 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.4 | 1.5 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.4 | 3.0 | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.4 | 11.1 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.4 | 4.4 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.4 | 6.0 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.3 | 7.5 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.3 | 3.4 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.3 | 33.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.3 | 1.3 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.3 | 4.5 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.3 | 1.3 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.3 | 3.1 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.3 | 2.7 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.3 | 1.5 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.3 | 5.8 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.3 | 2.5 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.3 | 2.7 | GO:0045329 | amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.3 | 0.8 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.3 | 3.7 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.3 | 3.7 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.3 | 3.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.2 | 0.7 | GO:0007500 | mesodermal cell fate determination(GO:0007500) negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.2 | 1.1 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.2 | 3.7 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.2 | 1.3 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.2 | 0.7 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.2 | 1.7 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.2 | 3.2 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.2 | 1.7 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) positive regulation of microtubule nucleation(GO:0090063) |
| 0.2 | 1.9 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.2 | 0.8 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.2 | 2.7 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.2 | 1.3 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.2 | 1.7 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.2 | 2.0 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.2 | 3.3 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.2 | 2.8 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.2 | 2.0 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.2 | 4.6 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.2 | 0.7 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) negative regulation of satellite cell differentiation(GO:1902725) |
| 0.2 | 0.5 | GO:1904387 | response to human chorionic gonadotropin(GO:0044752) cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.2 | 0.5 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.2 | 1.0 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.2 | 1.0 | GO:0060011 | progesterone biosynthetic process(GO:0006701) Sertoli cell proliferation(GO:0060011) |
| 0.2 | 2.0 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.2 | 2.0 | GO:0045794 | negative regulation of cell volume(GO:0045794) |
| 0.2 | 2.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 9.9 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.2 | 1.4 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.2 | 1.2 | GO:1900004 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.2 | 1.2 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.2 | 0.9 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.2 | 3.9 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.2 | 5.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 0.7 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 8.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.4 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.1 | 4.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 2.0 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 1.8 | GO:1902074 | response to salt(GO:1902074) |
| 0.1 | 0.8 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 1.6 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 3.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 1.6 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.5 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 1.3 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 2.9 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 2.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 6.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.5 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.1 | 0.6 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 1.0 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 0.4 | GO:1903011 | negative regulation of bone development(GO:1903011) |
| 0.1 | 0.8 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 10.1 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.1 | 1.3 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.1 | 2.4 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 6.7 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.1 | 0.8 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.0 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 1.1 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.1 | 1.9 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 1.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 1.5 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 0.3 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 4.0 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.1 | 2.5 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.1 | 0.6 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.1 | 0.8 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.9 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.1 | 2.0 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.1 | 0.6 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 | 1.5 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 1.2 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 2.2 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 2.6 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 1.9 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 2.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 1.9 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 1.0 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 5.6 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 1.0 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 | 1.0 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.1 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.1 | 1.4 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 4.1 | GO:0031640 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.1 | 5.4 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 1.0 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.1 | 1.0 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.1 | 0.5 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 1.3 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 0.4 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 | 1.9 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.1 | 1.4 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 4.8 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.1 | 4.2 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 0.8 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 2.0 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 7.7 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.1 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 2.3 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.1 | 0.5 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 1.6 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.1 | 1.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 5.0 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 0.9 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 0.9 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 1.4 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 1.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 2.3 | GO:0045840 | positive regulation of mitotic nuclear division(GO:0045840) |
| 0.1 | 1.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 2.4 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.6 | GO:0097283 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.0 | 0.1 | GO:0002034 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.0 | 0.7 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.9 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.2 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.8 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.7 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.0 | 0.8 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 1.3 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 3.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 4.7 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 1.1 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 2.6 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 1.0 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.1 | GO:0050718 | positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.0 | 4.9 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 2.1 | GO:0055007 | cardiac muscle cell differentiation(GO:0055007) |
| 0.0 | 0.1 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.0 | 0.5 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.0 | 0.5 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 1.0 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.5 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 1.5 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 11.6 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.7 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 4.5 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.5 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.2 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 1.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.3 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.7 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 2.3 | GO:0061351 | neural precursor cell proliferation(GO:0061351) |
| 0.0 | 1.2 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 1.8 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.3 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 0.0 | 0.5 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.0 | 0.1 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 1.0 | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway(GO:2001237) |
| 0.0 | 1.7 | GO:1904892 | regulation of JAK-STAT cascade(GO:0046425) regulation of STAT cascade(GO:1904892) |
| 0.0 | 0.7 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.5 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 1.1 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.7 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 0.3 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 0.6 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.1 | GO:0006702 | androgen biosynthetic process(GO:0006702) |
| 0.0 | 2.8 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 2.0 | GO:0072659 | protein localization to plasma membrane(GO:0072659) |
| 0.0 | 0.7 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.0 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.2 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 5.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 1.4 | 4.3 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 1.4 | 4.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 1.3 | 6.3 | GO:0044305 | calyx of Held(GO:0044305) |
| 1.2 | 3.6 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 1.1 | 14.0 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.9 | 18.0 | GO:0036038 | MKS complex(GO:0036038) |
| 0.8 | 16.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.7 | 2.2 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.7 | 3.3 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.6 | 4.8 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.5 | 8.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.5 | 1.6 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.5 | 2.6 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.5 | 3.9 | GO:0034464 | BBSome(GO:0034464) |
| 0.5 | 1.4 | GO:0033011 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
| 0.4 | 5.0 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.4 | 2.8 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.3 | 3.7 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.3 | 7.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.3 | 6.0 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.3 | 3.1 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.2 | 7.3 | GO:0005921 | gap junction(GO:0005921) |
| 0.2 | 2.3 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 6.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.2 | 1.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.2 | 1.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 2.6 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.2 | 1.3 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.2 | 8.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 1.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.2 | 1.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.2 | 0.5 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.2 | 1.2 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.2 | 6.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 5.3 | GO:0030990 | intraciliary transport particle(GO:0030990) |
| 0.2 | 1.0 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.2 | 2.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.2 | 1.7 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 10.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.5 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 1.0 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 5.6 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 3.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 8.1 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.1 | 3.4 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 1.6 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 1.5 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 2.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 15.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 12.6 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 1.6 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 4.2 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 13.1 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 2.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 5.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 4.4 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 2.5 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.6 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 11.4 | GO:0044439 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.1 | 6.0 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.1 | 2.6 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 40.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 0.8 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 1.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 3.3 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 4.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 1.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 1.0 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 4.5 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.1 | 0.5 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.1 | 0.3 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.1 | 2.3 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 1.6 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 2.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 1.0 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 1.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 2.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 6.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.9 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.9 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 6.1 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 1.0 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 5.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.9 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 1.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 7.6 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 5.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 5.1 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 4.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 8.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.9 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 2.8 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 1.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 1.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 3.8 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.9 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 53.8 | GO:0031224 | intrinsic component of membrane(GO:0031224) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 45.2 | GO:0008430 | selenium binding(GO:0008430) |
| 2.5 | 9.8 | GO:1904492 | Ac-Asp-Glu binding(GO:1904492) tetrahydrofolyl-poly(glutamate) polymer binding(GO:1904493) |
| 2.4 | 9.4 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 1.7 | 5.2 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 1.7 | 6.9 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) |
| 1.4 | 11.5 | GO:0019863 | IgE binding(GO:0019863) |
| 1.4 | 7.0 | GO:0008948 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) oxaloacetate decarboxylase activity(GO:0008948) |
| 1.4 | 4.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 1.3 | 5.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 1.2 | 6.0 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 1.0 | 6.3 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 1.0 | 3.0 | GO:0036505 | prosaposin receptor activity(GO:0036505) |
| 0.9 | 13.9 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.8 | 5.9 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.8 | 4.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.8 | 6.7 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.8 | 3.3 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.8 | 8.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.8 | 7.7 | GO:0005549 | odorant binding(GO:0005549) |
| 0.7 | 2.2 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.7 | 3.6 | GO:0018636 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) trans-1,2-dihydrobenzene-1,2-diol dehydrogenase activity(GO:0047115) |
| 0.7 | 15.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.6 | 1.7 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.6 | 2.8 | GO:0001016 | RNA polymerase III regulatory region DNA binding(GO:0001016) |
| 0.5 | 1.1 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.5 | 1.5 | GO:0042806 | fucose binding(GO:0042806) |
| 0.5 | 4.3 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.5 | 2.3 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.4 | 12.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.4 | 10.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.4 | 1.7 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.4 | 1.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.4 | 1.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.4 | 1.6 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.4 | 2.9 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.4 | 7.7 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.4 | 1.6 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.4 | 4.0 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.4 | 1.2 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.4 | 1.5 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.4 | 3.7 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.4 | 1.9 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.4 | 5.8 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.4 | 2.5 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.3 | 7.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.3 | 5.5 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.3 | 15.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.3 | 1.7 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.3 | 2.3 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.3 | 1.9 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.3 | 1.6 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.3 | 2.6 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.3 | 4.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.3 | 3.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.3 | 0.9 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.3 | 6.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.3 | 7.8 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.3 | 0.9 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.3 | 26.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.3 | 10.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.3 | 2.5 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.3 | 2.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.3 | 1.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.3 | 0.8 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.3 | 3.3 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.3 | 3.0 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.2 | 7.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.2 | 1.2 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.2 | 6.5 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.2 | 7.1 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 3.9 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.2 | 2.0 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.2 | 1.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 2.7 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.2 | 5.0 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 0.8 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.2 | 2.0 | GO:0015386 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 4.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 1.6 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 0.5 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.2 | 0.5 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.2 | 2.9 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 2.4 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.2 | 1.0 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.2 | 17.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 0.8 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.2 | 0.7 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.2 | 2.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.2 | 1.6 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.2 | 6.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.4 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.1 | 2.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 2.5 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.1 | 2.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.8 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 1.0 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 1.8 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.1 | 0.8 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 4.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 5.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 5.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 2.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.2 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 2.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 13.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 0.8 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 0.9 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 1.5 | GO:0042577 | phosphatidate phosphatase activity(GO:0008195) lipid phosphatase activity(GO:0042577) |
| 0.1 | 12.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 2.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 8.3 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 0.7 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 74.8 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 3.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 3.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 1.6 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 1.5 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 1.0 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 1.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.4 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 1.1 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 0.8 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 0.8 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 4.1 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.5 | GO:0019237 | satellite DNA binding(GO:0003696) centromeric DNA binding(GO:0019237) |
| 0.1 | 2.8 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 2.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 3.7 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 9.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 1.7 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 2.0 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 6.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 0.2 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 1.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.5 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 1.6 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.1 | 0.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 1.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 11.0 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 1.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 5.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.7 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 1.2 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 13.7 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 1.0 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 3.7 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 8.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 2.9 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 1.7 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.4 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 2.9 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.3 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 5.3 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.3 | GO:0008200 | ion channel inhibitor activity(GO:0008200) channel inhibitor activity(GO:0016248) |
| 0.0 | 0.9 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.1 | GO:0098960 | postsynaptic neurotransmitter receptor activity(GO:0098960) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 2.5 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.5 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 1.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 9.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.3 | 15.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.3 | 5.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 7.7 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.2 | 10.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 11.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 1.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 6.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 16.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 2.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 3.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 1.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 6.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 11.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 4.3 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 8.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 5.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 2.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 6.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 6.4 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 5.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.6 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 1.4 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 2.6 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 4.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 4.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 3.3 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.1 | 4.5 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 1.7 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 1.0 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 7.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 10.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.6 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 1.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 1.9 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 0.9 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 4.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 12.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 6.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 14.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.5 | 8.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.5 | 16.1 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.5 | 10.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.5 | 4.2 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.5 | 15.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.4 | 11.5 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.3 | 7.7 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.3 | 4.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.3 | 5.5 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.3 | 22.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 3.7 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 5.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 8.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.2 | 6.8 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 5.0 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.2 | 5.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 2.3 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.2 | 5.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 5.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 8.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.7 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 3.4 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 2.4 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.1 | 1.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 2.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.8 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 5.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 2.9 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 2.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 2.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 16.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 1.0 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 2.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 3.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.7 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.1 | 1.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 1.3 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 4.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 5.5 | REACTOME PI3K EVENTS IN ERBB4 SIGNALING | Genes involved in PI3K events in ERBB4 signaling |
| 0.1 | 3.0 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 1.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 5.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 2.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 2.0 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 1.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.9 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 8.2 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 1.6 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 1.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.6 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 1.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.8 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.0 | 0.6 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 2.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 1.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 2.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.5 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 2.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.8 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 1.2 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |