Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
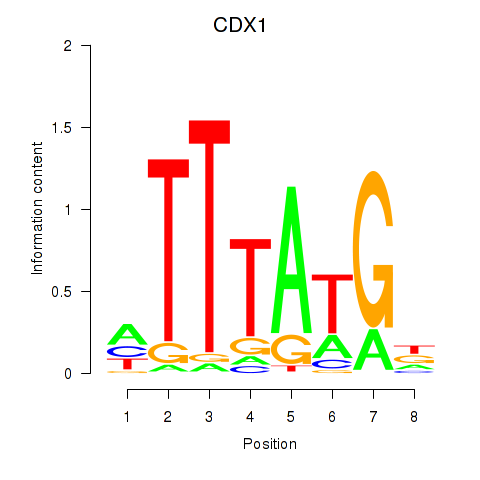
Results for CDX1
Z-value: 3.10
Transcription factors associated with CDX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CDX1
|
ENSG00000113722.17 | CDX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CDX1 | hg38_v1_chr5_+_150166770_150166788, hg38_v1_chr5_+_150166790_150166890 | 0.47 | 1.4e-13 | Click! |
Activity profile of CDX1 motif
Sorted Z-values of CDX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CDX1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 15.5 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 3.1 | 15.6 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 2.8 | 8.3 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 2.3 | 13.9 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 2.2 | 6.7 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 2.2 | 8.6 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 1.9 | 9.6 | GO:0072144 | mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) mesenchyme migration(GO:0090131) |
| 1.8 | 20.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 1.8 | 1.8 | GO:0051040 | regulation of calcium-independent cell-cell adhesion(GO:0051040) |
| 1.6 | 4.9 | GO:1904432 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 1.6 | 4.7 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 1.6 | 6.2 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 1.5 | 4.6 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 1.5 | 7.7 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 1.5 | 8.7 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 1.4 | 12.3 | GO:0072564 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 1.3 | 6.4 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 1.3 | 3.8 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 1.2 | 4.7 | GO:1900158 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 1.1 | 5.6 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 1.1 | 3.4 | GO:1903487 | regulation of lactation(GO:1903487) |
| 1.1 | 6.5 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 1.0 | 3.1 | GO:2001302 | cellular response to interleukin-13(GO:0035963) regulation of engulfment of apoptotic cell(GO:1901074) lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 1.0 | 3.1 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 1.0 | 13.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 1.0 | 2.9 | GO:0019605 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 0.9 | 2.8 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.9 | 3.8 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.9 | 3.8 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.9 | 2.8 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.9 | 21.7 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.9 | 5.2 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.9 | 3.5 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.9 | 12.8 | GO:0035878 | nail development(GO:0035878) |
| 0.8 | 5.9 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.8 | 8.4 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.8 | 4.5 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.7 | 3.6 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.7 | 3.6 | GO:1904501 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.7 | 10.1 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.7 | 4.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.7 | 2.6 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.6 | 6.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.6 | 4.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.6 | 2.3 | GO:0009698 | phenylpropanoid metabolic process(GO:0009698) coumarin metabolic process(GO:0009804) |
| 0.6 | 2.3 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.5 | 1.6 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.5 | 1.5 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.5 | 3.6 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.5 | 2.5 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.5 | 1.9 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.5 | 2.3 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.5 | 2.8 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.5 | 3.7 | GO:0002118 | aggressive behavior(GO:0002118) |
| 0.5 | 6.4 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.5 | 4.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.4 | 3.6 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.4 | 6.4 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.4 | 2.5 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.4 | 1.6 | GO:0019376 | galactolipid catabolic process(GO:0019376) |
| 0.4 | 1.9 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.4 | 2.2 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.4 | 1.8 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.4 | 2.5 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.3 | 1.7 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.3 | 6.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.3 | 1.6 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.3 | 3.5 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.3 | 4.7 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.3 | 1.5 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.3 | 8.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.3 | 0.6 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.3 | 3.5 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.3 | 1.3 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.3 | 2.0 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.3 | 1.5 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.2 | 0.7 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.2 | 0.5 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.2 | 1.2 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.2 | 1.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.2 | 3.6 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.2 | 4.1 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.2 | 4.3 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.2 | 0.6 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.2 | 1.4 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.2 | 5.7 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 3.4 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.2 | 2.6 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.2 | 4.3 | GO:0055069 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.2 | 3.7 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.2 | 1.0 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.2 | 3.6 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.2 | 0.5 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.2 | 3.8 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.2 | 1.6 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.2 | 1.0 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) stress-induced premature senescence(GO:0090400) |
| 0.2 | 2.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.2 | 4.2 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.2 | 6.9 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.2 | 1.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.2 | 0.6 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 3.9 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 3.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 5.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 2.1 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 8.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 13.7 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 23.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.4 | GO:0070846 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.1 | 0.3 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 2.9 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 2.2 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 1.3 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 5.2 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 2.2 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 1.0 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 12.0 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 1.1 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 4.6 | GO:0007099 | centriole replication(GO:0007099) |
| 0.1 | 0.7 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 1.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 1.4 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.1 | 1.4 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.1 | 0.7 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 3.1 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 0.8 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 0.5 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.1 | 5.0 | GO:0061028 | establishment of endothelial barrier(GO:0061028) |
| 0.1 | 0.5 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 1.7 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.1 | 0.6 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 2.6 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.1 | 0.3 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 | 2.5 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.1 | 2.7 | GO:0040018 | positive regulation of multicellular organism growth(GO:0040018) |
| 0.1 | 4.9 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.1 | 3.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 1.0 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.1 | 1.8 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 5.3 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 0.8 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.1 | 0.5 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 1.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 0.4 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 1.3 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 2.0 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 2.6 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 3.2 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 1.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 3.1 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.4 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 1.6 | GO:2000273 | positive regulation of receptor activity(GO:2000273) |
| 0.0 | 1.4 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 2.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 2.1 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 1.6 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.7 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.5 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 1.0 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.5 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.0 | 1.3 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 1.6 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 1.5 | GO:0061053 | somite development(GO:0061053) |
| 0.0 | 0.3 | GO:0034391 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 | 2.5 | GO:0055001 | muscle cell development(GO:0055001) |
| 0.0 | 0.8 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 13.9 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 3.2 | 9.6 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 1.3 | 20.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 1.1 | 3.2 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 1.0 | 13.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.8 | 2.5 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.8 | 4.9 | GO:1990357 | terminal web(GO:1990357) |
| 0.8 | 4.6 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.8 | 3.8 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.8 | 6.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.7 | 3.7 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.7 | 7.7 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.7 | 4.7 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.6 | 7.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.6 | 5.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.6 | 5.0 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.6 | 3.6 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.6 | 4.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.6 | 3.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.5 | 8.6 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.5 | 7.6 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.5 | 1.5 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.4 | 5.8 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.4 | 17.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.4 | 14.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.3 | 13.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.3 | 3.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 13.2 | GO:0031430 | M band(GO:0031430) |
| 0.3 | 5.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.2 | 1.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 20.5 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.2 | 4.1 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 0.6 | GO:0071756 | dimeric IgA immunoglobulin complex(GO:0071750) secretory dimeric IgA immunoglobulin complex(GO:0071752) IgM immunoglobulin complex(GO:0071753) IgM immunoglobulin complex, circulating(GO:0071754) pentameric IgM immunoglobulin complex(GO:0071756) |
| 0.2 | 2.0 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.2 | 1.0 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.2 | 5.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 1.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.2 | 3.5 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.2 | 2.1 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.2 | 3.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 6.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 1.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 3.0 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 2.6 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.1 | 3.6 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 10.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 6.4 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 4.8 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 2.3 | GO:0036019 | endolysosome(GO:0036019) endolysosome membrane(GO:0036020) |
| 0.1 | 15.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 3.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 7.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 4.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 7.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 2.0 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 2.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 1.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 3.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 3.1 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 2.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 8.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 5.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 6.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 61.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 2.2 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.3 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.5 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 5.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 2.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.4 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 2.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.7 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.8 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 2.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.9 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.2 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 1.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 23.9 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 2.2 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 15.5 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 2.2 | 6.7 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 2.2 | 6.5 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 1.6 | 6.4 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 1.6 | 6.2 | GO:0035276 | aldehyde oxidase activity(GO:0004031) ethanol binding(GO:0035276) |
| 1.5 | 12.3 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 1.5 | 4.5 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 1.4 | 15.6 | GO:0031433 | telethonin binding(GO:0031433) |
| 1.2 | 3.7 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 1.2 | 14.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 1.2 | 4.7 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 1.0 | 4.2 | GO:0035473 | lipase binding(GO:0035473) |
| 1.0 | 3.1 | GO:0047977 | hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 1.0 | 4.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 1.0 | 3.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 1.0 | 3.9 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 1.0 | 20.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.9 | 2.8 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.9 | 3.6 | GO:0052596 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.9 | 6.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.8 | 2.5 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.8 | 8.2 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.7 | 3.6 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.7 | 2.7 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.7 | 4.7 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.7 | 2.6 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.6 | 3.8 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.6 | 6.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.6 | 5.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.6 | 6.7 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.5 | 8.6 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.5 | 1.6 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.5 | 2.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.5 | 7.9 | GO:0016918 | retinal binding(GO:0016918) |
| 0.5 | 3.7 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.5 | 1.4 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.4 | 8.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.4 | 2.0 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.4 | 2.0 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.4 | 2.3 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.4 | 1.9 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.4 | 2.9 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.4 | 8.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.4 | 2.5 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.3 | 2.0 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.3 | 1.6 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.3 | 6.1 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.3 | 3.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.3 | 4.9 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.3 | 2.9 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.3 | 2.0 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 8.0 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.3 | 1.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.3 | 2.2 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.3 | 7.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 4.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.3 | 2.3 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.2 | 2.2 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.2 | 0.7 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.2 | 4.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 1.6 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.2 | 5.4 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 25.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 3.6 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.2 | 3.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.2 | 3.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 4.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 9.1 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.2 | 1.0 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.2 | 2.6 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.2 | 2.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 1.7 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.2 | 3.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 5.0 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.2 | 5.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 5.9 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 1.8 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.1 | 3.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 16.6 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 0.6 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 3.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 10.6 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 1.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 5.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 1.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 3.5 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 10.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.5 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 0.8 | GO:0030883 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.1 | 1.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 25.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 2.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 1.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 1.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 5.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 2.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 4.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 1.7 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 0.5 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.7 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 2.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 2.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 2.0 | GO:0042166 | acetylcholine-gated cation channel activity(GO:0022848) acetylcholine binding(GO:0042166) |
| 0.1 | 0.3 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 1.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 1.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 1.0 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.8 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) lipid phosphatase activity(GO:0042577) |
| 0.1 | 1.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.4 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 1.4 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 35.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 1.0 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 1.0 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.3 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 2.7 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.6 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 2.9 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 2.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.7 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 2.7 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 0.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.5 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.3 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) TFIIB-class transcription factor binding(GO:0001093) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 9.3 | GO:0008270 | zinc ion binding(GO:0008270) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 20.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.6 | 28.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 13.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 4.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 6.0 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 11.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 9.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 3.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 9.8 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 2.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 3.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 20.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 0.7 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 3.7 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 8.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 3.0 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 3.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 5.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 3.2 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.1 | 1.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 1.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 1.7 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 2.9 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 3.2 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 2.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 2.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.4 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 12.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.6 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 3.5 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.7 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 1.3 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 7.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 9.8 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.6 | 15.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.6 | 5.6 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.5 | 13.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.5 | 7.9 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.5 | 6.7 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.4 | 2.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.4 | 37.7 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.3 | 6.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.3 | 12.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.3 | 13.9 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.3 | 7.2 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.3 | 5.2 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.3 | 7.9 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.3 | 4.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 2.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.2 | 3.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 4.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 3.1 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 19.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 2.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 3.0 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 2.6 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 4.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 6.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 4.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 4.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 10.5 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 2.0 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 4.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 3.2 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 1.9 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.1 | 3.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 3.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 9.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 2.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 1.7 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 3.4 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 10.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 6.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.0 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.1 | 2.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 2.2 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 1.0 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 1.5 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 3.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.4 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 2.4 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.0 | 1.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 4.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.7 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 2.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 2.9 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 1.4 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 1.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.2 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 1.8 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |