Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
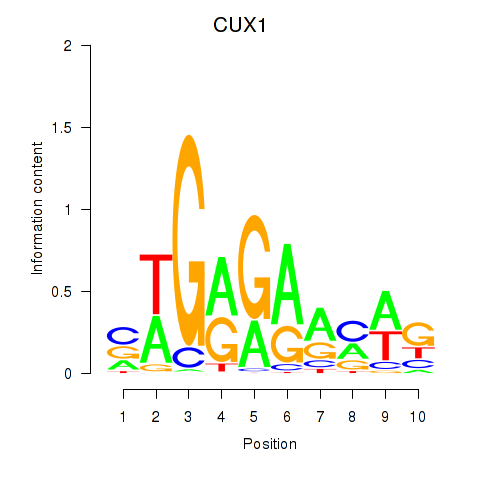
Results for CUX1
Z-value: 11.55
Transcription factors associated with CUX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CUX1
|
ENSG00000257923.12 | CUX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CUX1 | hg38_v1_chr7_+_101815983_101816030 | 0.34 | 2.0e-07 | Click! |
Activity profile of CUX1 motif
Sorted Z-values of CUX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CUX1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.6 | 49.8 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 15.0 | 45.1 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 8.6 | 25.8 | GO:0051941 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 7.9 | 23.8 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 7.1 | 28.5 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 6.9 | 20.6 | GO:0001983 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 6.8 | 27.1 | GO:1904980 | positive regulation of endosome organization(GO:1904980) |
| 6.2 | 18.5 | GO:1900238 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 5.6 | 16.8 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 5.3 | 15.8 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 5.2 | 21.0 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 4.4 | 17.5 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 4.2 | 16.6 | GO:1903611 | regulation of calcium-dependent ATPase activity(GO:1903610) negative regulation of calcium-dependent ATPase activity(GO:1903611) regulation of dUTP diphosphatase activity(GO:1903627) positive regulation of dUTP diphosphatase activity(GO:1903629) negative regulation of aminoacyl-tRNA ligase activity(GO:1903631) regulation of leucine-tRNA ligase activity(GO:1903633) negative regulation of leucine-tRNA ligase activity(GO:1903634) |
| 4.0 | 27.8 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 3.6 | 21.7 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 3.5 | 14.0 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 3.4 | 10.3 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 3.3 | 9.9 | GO:0001794 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) |
| 3.2 | 12.9 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 3.2 | 12.8 | GO:0050904 | diapedesis(GO:0050904) glomerular endothelium development(GO:0072011) |
| 3.0 | 8.9 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 2.8 | 19.4 | GO:0071279 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) cellular response to cobalt ion(GO:0071279) |
| 2.7 | 8.1 | GO:2000537 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 2.6 | 13.2 | GO:0072708 | response to sorbitol(GO:0072708) |
| 2.6 | 7.8 | GO:1903937 | response to acrylamide(GO:1903937) |
| 2.5 | 2.5 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 2.5 | 9.9 | GO:0035054 | atrial septum secundum morphogenesis(GO:0003290) embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 2.3 | 9.4 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 2.2 | 13.2 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 2.2 | 36.7 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 2.2 | 15.1 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 2.1 | 6.4 | GO:0099557 | trans-synaptic signaling by trans-synaptic complex, modulating synaptic transmission(GO:0099557) |
| 2.1 | 8.3 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 2.1 | 10.4 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 2.1 | 8.2 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 2.0 | 6.0 | GO:0010958 | regulation of amino acid import(GO:0010958) |
| 1.9 | 5.7 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 1.9 | 20.5 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 1.8 | 12.9 | GO:0080009 | mRNA methylation(GO:0080009) |
| 1.8 | 10.9 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 1.8 | 5.4 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 1.8 | 12.4 | GO:1990822 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 1.8 | 22.9 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 1.7 | 19.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 1.7 | 15.2 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 1.7 | 8.4 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 1.7 | 5.0 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 1.7 | 16.6 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 1.6 | 6.4 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 1.6 | 8.0 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 1.6 | 4.8 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 1.6 | 1.6 | GO:0060459 | left lung development(GO:0060459) |
| 1.6 | 14.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 1.4 | 4.3 | GO:0019075 | virus maturation(GO:0019075) |
| 1.4 | 2.9 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 1.4 | 8.6 | GO:0061511 | centriole elongation(GO:0061511) |
| 1.4 | 5.7 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 1.4 | 8.4 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 1.4 | 4.1 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 1.4 | 16.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 1.4 | 12.4 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 1.3 | 4.0 | GO:1990451 | cellular stress response to acidic pH(GO:1990451) |
| 1.3 | 10.7 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 1.3 | 5.3 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 1.3 | 4.0 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 1.3 | 10.4 | GO:0061709 | reticulophagy(GO:0061709) |
| 1.3 | 6.4 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 1.3 | 10.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 1.3 | 3.8 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 1.2 | 16.2 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 1.2 | 7.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 1.2 | 84.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 1.2 | 4.6 | GO:0072134 | hyaluranon cable assembly(GO:0036118) nephrogenic mesenchyme morphogenesis(GO:0072134) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) allantois development(GO:1905069) |
| 1.1 | 3.4 | GO:0060032 | notochord regression(GO:0060032) |
| 1.1 | 7.9 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 1.1 | 16.5 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 1.1 | 4.2 | GO:0010193 | response to ozone(GO:0010193) |
| 1.0 | 6.0 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 1.0 | 5.8 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 1.0 | 44.3 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 1.0 | 3.8 | GO:1904640 | response to methionine(GO:1904640) |
| 1.0 | 7.6 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.9 | 4.6 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.9 | 1.8 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.9 | 5.4 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.9 | 3.6 | GO:0002331 | pre-B cell differentiation(GO:0002329) pre-B cell allelic exclusion(GO:0002331) |
| 0.9 | 6.2 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.9 | 17.6 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.9 | 7.0 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.9 | 3.5 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.9 | 10.4 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.9 | 4.3 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.9 | 1.7 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.9 | 6.8 | GO:0086043 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.8 | 2.5 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.8 | 5.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.8 | 7.4 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.8 | 3.3 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.8 | 10.6 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.8 | 2.4 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.8 | 6.5 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.8 | 2.4 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.8 | 4.7 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.8 | 5.5 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.8 | 3.8 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.8 | 3.0 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.8 | 3.8 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.7 | 4.5 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.7 | 1.5 | GO:0009624 | response to nematode(GO:0009624) |
| 0.7 | 3.7 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.7 | 2.2 | GO:0018874 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 0.7 | 2.1 | GO:0018879 | biphenyl metabolic process(GO:0018879) dibenzo-p-dioxin metabolic process(GO:0018894) phthalate metabolic process(GO:0018963) cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.7 | 4.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.7 | 8.3 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.7 | 7.6 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.7 | 4.8 | GO:0001999 | renal response to blood flow involved in circulatory renin-angiotensin regulation of systemic arterial blood pressure(GO:0001999) renin secretion into blood stream(GO:0002001) |
| 0.7 | 6.8 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.7 | 2.0 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) |
| 0.7 | 4.7 | GO:0042423 | catechol-containing compound biosynthetic process(GO:0009713) catecholamine biosynthetic process(GO:0042423) |
| 0.7 | 25.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.7 | 6.5 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.7 | 5.9 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.6 | 1.9 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.6 | 6.9 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.6 | 3.1 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.6 | 3.1 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.6 | 11.8 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.6 | 7.5 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.6 | 6.1 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.6 | 3.1 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.6 | 8.5 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.6 | 2.4 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.6 | 6.5 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.6 | 7.7 | GO:0038129 | ERBB3 signaling pathway(GO:0038129) |
| 0.6 | 2.4 | GO:0019376 | galactolipid catabolic process(GO:0019376) |
| 0.6 | 2.3 | GO:0050893 | sensory processing(GO:0050893) |
| 0.6 | 20.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.6 | 51.7 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.6 | 5.2 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.6 | 10.3 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.6 | 4.0 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.6 | 3.4 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.5 | 7.1 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.5 | 16.2 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.5 | 1.6 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.5 | 5.4 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.5 | 13.2 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.5 | 29.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.5 | 3.1 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.5 | 11.6 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.5 | 43.0 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.5 | 4.5 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.5 | 3.3 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.5 | 2.3 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.5 | 1.8 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.5 | 3.7 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.5 | 1.8 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.5 | 42.4 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.4 | 18.4 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.4 | 2.2 | GO:0045416 | positive regulation of interleukin-8 biosynthetic process(GO:0045416) |
| 0.4 | 2.1 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.4 | 3.4 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.4 | 11.4 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.4 | 5.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.4 | 12.5 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.4 | 6.3 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.4 | 6.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.4 | 11.5 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.4 | 4.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.4 | 4.4 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.4 | 0.8 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.4 | 37.4 | GO:0032945 | negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) |
| 0.4 | 1.9 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.4 | 5.8 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.4 | 17.0 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.4 | 0.7 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.4 | 3.7 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.4 | 3.7 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.4 | 7.2 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.4 | 3.6 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.4 | 17.0 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.4 | 1.8 | GO:0010816 | neuropeptide catabolic process(GO:0010813) substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.3 | 0.3 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.3 | 23.1 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.3 | 1.4 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.3 | 10.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.3 | 51.1 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.3 | 26.4 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.3 | 1.0 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.3 | 2.7 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.3 | 2.6 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.3 | 14.6 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.3 | 2.5 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.3 | 4.7 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.3 | 2.5 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.3 | 14.5 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.3 | 0.9 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.3 | 4.2 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.3 | 4.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.3 | 9.7 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.3 | 1.7 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.3 | 8.6 | GO:0097503 | sialylation(GO:0097503) |
| 0.3 | 5.1 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.3 | 8.6 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.3 | 3.0 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.3 | 2.5 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.3 | 19.2 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.3 | 1.9 | GO:0086100 | endothelin receptor signaling pathway(GO:0086100) |
| 0.3 | 4.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.3 | 4.0 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.3 | 3.4 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.3 | 1.0 | GO:1904382 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) glycoprotein ERAD pathway(GO:0097466) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.3 | 8.1 | GO:0000732 | strand displacement(GO:0000732) |
| 0.3 | 9.9 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.3 | 11.5 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.2 | 1.5 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.2 | 1.5 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.2 | 2.1 | GO:0035860 | negative regulation of protein autophosphorylation(GO:0031953) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.2 | 6.5 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.2 | 1.6 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.2 | 5.7 | GO:0051181 | cofactor transport(GO:0051181) |
| 0.2 | 3.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.2 | 9.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.2 | 3.6 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.2 | 20.8 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.2 | 3.6 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.2 | 6.3 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.2 | 4.2 | GO:0030575 | nuclear body organization(GO:0030575) positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 1.2 | GO:0046618 | drug export(GO:0046618) |
| 0.2 | 7.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.2 | 1.9 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.2 | 0.7 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.2 | 2.4 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.2 | 0.7 | GO:0032627 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) positive regulation of interleukin-23 production(GO:0032747) |
| 0.2 | 4.3 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.2 | 12.0 | GO:0032642 | regulation of chemokine production(GO:0032642) |
| 0.2 | 3.7 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.2 | 9.4 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.2 | 0.5 | GO:0089700 | protein kinase D signaling(GO:0089700) positive regulation of deacetylase activity(GO:0090045) positive regulation of histone deacetylase activity(GO:1901727) |
| 0.2 | 3.3 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.2 | 3.0 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 1.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 6.9 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.2 | 6.7 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.2 | 1.7 | GO:0007620 | insemination(GO:0007320) copulation(GO:0007620) |
| 0.2 | 4.4 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.2 | 1.6 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.2 | 6.9 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.2 | 2.3 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.2 | 2.6 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.1 | 0.3 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 3.6 | GO:0040018 | positive regulation of multicellular organism growth(GO:0040018) |
| 0.1 | 1.6 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.3 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 4.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 1.1 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 6.0 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 1.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.1 | 7.4 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.1 | 1.0 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 2.0 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 2.8 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 | 4.6 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.1 | 1.2 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 6.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 6.5 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 0.7 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.1 | 3.3 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 | 4.6 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 3.0 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 1.0 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 4.7 | GO:0000060 | protein import into nucleus, translocation(GO:0000060) |
| 0.1 | 1.3 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.1 | 0.4 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.1 | 7.9 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 5.7 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 5.1 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.1 | 6.3 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 | 5.4 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.1 | 8.0 | GO:0042633 | molting cycle(GO:0042303) hair cycle(GO:0042633) |
| 0.1 | 1.8 | GO:0035590 | response to ATP(GO:0033198) purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.1 | 0.2 | GO:0003418 | growth plate cartilage chondrocyte differentiation(GO:0003418) chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.1 | 3.0 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 1.1 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 4.9 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 2.6 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 27.8 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.1 | 0.7 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.1 | 11.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.8 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.1 | 1.0 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 2.9 | GO:0002228 | natural killer cell mediated immunity(GO:0002228) |
| 0.1 | 9.7 | GO:0030518 | intracellular steroid hormone receptor signaling pathway(GO:0030518) |
| 0.1 | 4.5 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.1 | 0.5 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 0.7 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 3.4 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 8.4 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 2.5 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.2 | GO:0003360 | brainstem development(GO:0003360) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 2.6 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 4.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.5 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.2 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 1.2 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.9 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 2.4 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 1.4 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 2.2 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 2.5 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 2.6 | GO:0015992 | proton transport(GO:0015992) |
| 0.0 | 0.6 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 3.0 | GO:0045665 | negative regulation of neuron differentiation(GO:0045665) |
| 0.0 | 4.7 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.3 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 1.2 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 1.5 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.4 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.9 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.4 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.8 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 0.9 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.3 | 45.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 3.1 | 9.2 | GO:0071756 | IgM immunoglobulin complex(GO:0071753) IgM immunoglobulin complex, circulating(GO:0071754) pentameric IgM immunoglobulin complex(GO:0071756) |
| 2.5 | 10.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 2.3 | 25.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 2.2 | 8.6 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 2.0 | 7.8 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 1.8 | 19.4 | GO:0043203 | axon hillock(GO:0043203) |
| 1.7 | 14.0 | GO:0035976 | AP1 complex(GO:0035976) |
| 1.7 | 10.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 1.7 | 84.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 1.7 | 32.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 1.6 | 8.0 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 1.5 | 19.4 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 1.4 | 25.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 1.3 | 10.3 | GO:0060199 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 1.2 | 8.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 1.2 | 19.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 1.1 | 6.8 | GO:0071439 | clathrin complex(GO:0071439) |
| 1.1 | 16.8 | GO:0097433 | dense body(GO:0097433) |
| 1.1 | 11.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 1.1 | 15.8 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 1.0 | 9.4 | GO:0032010 | phagolysosome(GO:0032010) |
| 1.0 | 16.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 1.0 | 4.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 1.0 | 8.7 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.9 | 2.7 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.8 | 2.5 | GO:0055087 | Ski complex(GO:0055087) |
| 0.8 | 8.4 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.8 | 7.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.8 | 5.5 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.8 | 27.1 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.8 | 8.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.7 | 9.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.7 | 4.2 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.7 | 4.7 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.7 | 13.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.6 | 3.8 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.6 | 1.8 | GO:0033011 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
| 0.6 | 3.7 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.6 | 15.1 | GO:0031095 | spectrin-associated cytoskeleton(GO:0014731) platelet dense tubular network membrane(GO:0031095) |
| 0.6 | 3.6 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.6 | 24.4 | GO:0031430 | M band(GO:0031430) |
| 0.6 | 3.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.5 | 1.6 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.5 | 26.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.5 | 3.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.5 | 58.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.5 | 2.5 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.5 | 6.9 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.5 | 5.8 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.5 | 3.9 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.5 | 4.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.4 | 0.9 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.4 | 4.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.4 | 8.1 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.4 | 4.6 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.4 | 2.5 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.4 | 14.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.4 | 5.4 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.4 | 43.9 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.4 | 30.1 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.4 | 8.6 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.4 | 7.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.4 | 3.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.4 | 1.5 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.4 | 15.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.4 | 1.8 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.4 | 1.8 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.4 | 6.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.3 | 4.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.3 | 29.7 | GO:0005776 | autophagosome(GO:0005776) |
| 0.3 | 8.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.3 | 2.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.3 | 11.9 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.3 | 8.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.3 | 7.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.3 | 11.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.3 | 21.3 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.3 | 51.1 | GO:0030426 | growth cone(GO:0030426) |
| 0.3 | 5.0 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.3 | 2.6 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.3 | 15.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 23.7 | GO:0016234 | inclusion body(GO:0016234) |
| 0.2 | 12.1 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.2 | 2.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.2 | 8.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 9.3 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.2 | 7.4 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.2 | 5.5 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.2 | 6.9 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.2 | 14.0 | GO:0005902 | microvillus(GO:0005902) |
| 0.2 | 2.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.2 | 3.4 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.2 | 21.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 3.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 10.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.2 | 6.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.2 | 2.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.2 | 39.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.2 | 41.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.2 | 13.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.2 | 8.1 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.2 | 13.5 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.2 | 1.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.2 | 7.2 | GO:0097546 | ciliary base(GO:0097546) |
| 0.2 | 5.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 4.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 3.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 5.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 11.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 2.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 167.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 3.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 2.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 9.3 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 19.9 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 11.4 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 8.9 | GO:0098793 | presynapse(GO:0098793) |
| 0.1 | 1.0 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 2.0 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 1.0 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 5.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 3.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 13.6 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.1 | 3.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 9.8 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 4.8 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.1 | 19.6 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 1.3 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.7 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 3.0 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.7 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 10.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.1 | GO:0098533 | ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.0 | 2.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 3.5 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.6 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.9 | GO:0032587 | ruffle membrane(GO:0032587) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.0 | 23.9 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 6.9 | 27.8 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 6.8 | 33.9 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 6.4 | 45.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 6.3 | 25.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 6.2 | 18.5 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 4.3 | 12.9 | GO:0008988 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 4.2 | 16.6 | GO:0004341 | gluconolactonase activity(GO:0004341) |
| 3.9 | 27.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 3.6 | 14.6 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 3.6 | 21.7 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 3.2 | 12.9 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 3.1 | 12.4 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 2.8 | 19.7 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 2.7 | 8.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 2.5 | 10.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 2.4 | 7.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 2.3 | 35.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 2.3 | 2.3 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 2.3 | 6.9 | GO:0004040 | amidase activity(GO:0004040) |
| 2.3 | 6.9 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 2.2 | 8.9 | GO:0052596 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 2.2 | 13.2 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 2.1 | 8.5 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 2.1 | 8.4 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 2.1 | 10.3 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 2.1 | 8.2 | GO:0035276 | aldehyde oxidase activity(GO:0004031) ethanol binding(GO:0035276) |
| 2.0 | 6.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 2.0 | 25.8 | GO:1990239 | sodium:potassium-exchanging ATPase activity(GO:0005391) steroid hormone binding(GO:1990239) |
| 2.0 | 5.9 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 2.0 | 29.7 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 1.9 | 5.7 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 1.8 | 32.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 1.8 | 7.2 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 1.8 | 5.3 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 1.8 | 7.0 | GO:0050473 | arachidonate 15-lipoxygenase activity(GO:0050473) |
| 1.7 | 3.3 | GO:0042806 | fucose binding(GO:0042806) |
| 1.6 | 6.5 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 1.6 | 4.8 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 1.6 | 4.7 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 1.5 | 89.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 1.3 | 4.0 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 1.3 | 17.5 | GO:0009374 | biotin binding(GO:0009374) |
| 1.2 | 7.4 | GO:0051373 | FATZ binding(GO:0051373) |
| 1.2 | 3.6 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 1.2 | 4.7 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 1.2 | 9.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 1.1 | 3.4 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 1.1 | 4.5 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) |
| 1.1 | 7.9 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 1.1 | 7.6 | GO:0005499 | vitamin D binding(GO:0005499) |
| 1.1 | 14.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 1.0 | 12.4 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 1.0 | 3.1 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 1.0 | 19.3 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 1.0 | 4.9 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 1.0 | 2.9 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.9 | 8.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.9 | 19.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.9 | 25.5 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.9 | 3.6 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.9 | 9.9 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.9 | 3.5 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.9 | 3.4 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.8 | 4.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.8 | 7.6 | GO:0030883 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.8 | 2.5 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.8 | 29.3 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.8 | 17.0 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.8 | 8.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.8 | 4.0 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.8 | 6.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.8 | 24.3 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.8 | 6.3 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.8 | 9.4 | GO:0030911 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) TPR domain binding(GO:0030911) |
| 0.8 | 4.7 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.8 | 3.0 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.7 | 5.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.7 | 4.4 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.7 | 5.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.7 | 13.2 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.7 | 4.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.7 | 2.0 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.7 | 5.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.7 | 6.5 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.6 | 11.0 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.6 | 1.9 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.6 | 18.3 | GO:0071949 | FAD binding(GO:0071949) |
| 0.6 | 7.4 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.6 | 5.5 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.6 | 11.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.6 | 6.5 | GO:0004568 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.6 | 3.5 | GO:0023029 | MHC class Ib protein binding(GO:0023029) |
| 0.6 | 12.3 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.6 | 20.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.6 | 9.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.6 | 11.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.6 | 158.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.6 | 61.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.5 | 3.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.5 | 9.5 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.5 | 21.0 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.5 | 7.8 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.5 | 4.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.5 | 4.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.5 | 31.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.5 | 15.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.5 | 5.4 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.5 | 6.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.5 | 3.8 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.5 | 1.4 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.5 | 4.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.5 | 3.7 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.5 | 5.5 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.4 | 6.0 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.4 | 6.2 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.4 | 13.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.4 | 4.7 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.4 | 36.1 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.4 | 2.7 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.4 | 1.5 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.4 | 3.0 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.4 | 4.5 | GO:0008430 | selenium binding(GO:0008430) |
| 0.4 | 11.9 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.4 | 4.6 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.4 | 2.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.3 | 3.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.3 | 5.8 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.3 | 13.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.3 | 1.3 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.3 | 5.6 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.3 | 4.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.3 | 27.9 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.3 | 7.5 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.3 | 3.4 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.3 | 6.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.3 | 2.1 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.3 | 1.2 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.3 | 11.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.3 | 10.0 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.3 | 1.2 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 0.3 | 7.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.3 | 1.7 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) arginine binding(GO:0034618) |
| 0.3 | 3.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.3 | 9.4 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.3 | 3.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.3 | 11.5 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.3 | 7.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.3 | 1.9 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.3 | 2.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.3 | 47.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.3 | 5.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 2.7 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.3 | 0.8 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.3 | 5.0 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.3 | 4.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.3 | 1.6 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.3 | 19.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 1.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.2 | 15.1 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.2 | 1.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.2 | 6.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 2.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.2 | 6.3 | GO:0008066 | glutamate receptor activity(GO:0008066) |
| 0.2 | 4.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 4.0 | GO:0042379 | chemokine receptor binding(GO:0042379) |
| 0.2 | 0.4 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.2 | 2.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.2 | 8.5 | GO:0004601 | peroxidase activity(GO:0004601) oxidoreductase activity, acting on peroxide as acceptor(GO:0016684) |
| 0.2 | 2.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 3.4 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.2 | 12.3 | GO:0004402 | histone acetyltransferase activity(GO:0004402) peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.2 | 2.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 4.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 1.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.2 | 4.6 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.2 | 6.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 2.7 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.2 | 2.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.2 | 8.9 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.2 | 2.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 2.5 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.2 | 1.6 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.2 | 4.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 15.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.2 | 6.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 3.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.2 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.1 | 10.6 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.1 | 3.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.4 | GO:0004629 | phospholipase C activity(GO:0004629) |
| 0.1 | 11.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 2.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 1.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 13.6 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 7.7 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 1.0 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 6.9 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 1.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 10.1 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 8.3 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 3.0 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 3.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 10.5 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) |
| 0.1 | 4.0 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 4.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 8.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 2.1 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.1 | 2.9 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 3.6 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 1.4 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 2.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 3.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 1.0 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 1.0 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 10.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 16.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 3.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 3.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.4 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 1.6 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 0.2 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 3.4 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 2.2 | GO:0008200 | ion channel inhibitor activity(GO:0008200) channel inhibitor activity(GO:0016248) |
| 0.0 | 0.4 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 1.0 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 1.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 1.3 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.7 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 5.7 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.9 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.8 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 1.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.9 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 17.1 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 3.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 76.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 1.1 | 19.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.8 | 16.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.6 | 13.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.5 | 6.5 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.5 | 16.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.4 | 8.9 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.4 | 11.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.4 | 14.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.4 | 15.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.4 | 62.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.4 | 34.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.4 | 21.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.4 | 25.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.3 | 6.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.3 | 10.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.3 | 14.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.3 | 4.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.3 | 9.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.3 | 74.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 4.2 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.2 | 12.0 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.2 | 1.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 13.0 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.2 | 6.4 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.2 | 9.3 | PID ATM PATHWAY | ATM pathway |
| 0.2 | 2.6 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.2 | 7.1 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.2 | 3.7 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.2 | 2.6 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.2 | 14.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.2 | 2.0 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 62.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.2 | 6.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.2 | 9.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.2 | 7.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 11.9 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.2 | 9.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.2 | 11.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.2 | 9.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 1.9 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 5.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 7.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 16.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 5.3 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 5.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 4.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 1.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 4.9 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 3.3 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 9.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 2.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 4.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 15.1 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.1 | 5.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 1.0 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 2.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 1.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 2.2 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.5 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.9 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.0 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 54.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 2.1 | 49.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 1.6 | 26.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 1.2 | 8.5 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 1.2 | 17.4 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 1.1 | 37.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 1.1 | 19.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 1.0 | 18.7 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 1.0 | 27.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 1.0 | 10.9 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.9 | 6.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.9 | 6.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.9 | 21.4 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.8 | 8.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.8 | 16.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.8 | 15.8 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.8 | 10.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.7 | 20.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.7 | 20.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.7 | 13.0 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.7 | 45.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.6 | 15.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.6 | 25.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.6 | 11.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.5 | 25.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.5 | 3.7 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.5 | 14.4 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.4 | 14.1 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.4 | 4.7 | REACTOME OPSINS | Genes involved in Opsins |
| 0.4 | 17.8 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.4 | 7.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.3 | 6.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.3 | 36.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.3 | 15.0 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.3 | 20.5 | REACTOME IL 2 SIGNALING | Genes involved in Interleukin-2 signaling |
| 0.3 | 23.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.3 | 15.6 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.3 | 6.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 23.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.3 | 16.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.3 | 9.9 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.3 | 5.6 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.3 | 5.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.3 | 3.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.3 | 5.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.3 | 5.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.3 | 6.5 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.2 | 5.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 4.9 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.2 | 3.7 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.2 | 11.1 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.2 | 8.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.2 | 16.9 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.2 | 5.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 19.3 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 28.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.2 | 4.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.2 | 4.9 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.2 | 5.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 5.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.2 | 3.4 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.2 | 4.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 7.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 13.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.2 | 10.2 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.2 | 5.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 13.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.2 | 8.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 0.9 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 20.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 3.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 2.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 3.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 5.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 6.5 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 7.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 5.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 3.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 2.3 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.1 | 6.7 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.1 | 3.5 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 0.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 1.0 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 4.8 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.1 | 1.0 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 3.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.7 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 2.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 2.8 | REACTOME SIGNALLING BY NGF | Genes involved in Signalling by NGF |
| 0.0 | 1.2 | REACTOME GPCR LIGAND BINDING | Genes involved in GPCR ligand binding |
| 0.0 | 0.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |