Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for DBX2_HLX
Z-value: 1.95
Transcription factors associated with DBX2_HLX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
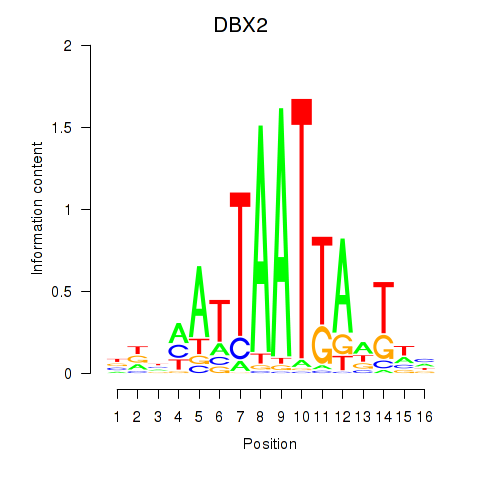
DBX2
|
ENSG00000185610.6 | DBX2 |
|
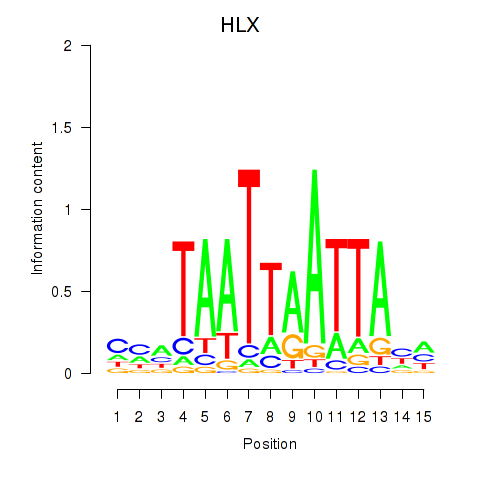
HLX
|
ENSG00000136630.13 | HLX |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HLX | hg38_v1_chr1_+_220879434_220879457 | -0.03 | 6.2e-01 | Click! |
Activity profile of DBX2_HLX motif
Sorted Z-values of DBX2_HLX motif
Network of associatons between targets according to the STRING database.
First level regulatory network of DBX2_HLX
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 13.3 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 2.6 | 7.7 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 2.2 | 6.6 | GO:0036292 | DNA rewinding(GO:0036292) |
| 2.1 | 6.2 | GO:0034092 | negative regulation of maintenance of sister chromatid cohesion(GO:0034092) negative regulation of maintenance of mitotic sister chromatid cohesion(GO:0034183) maintenance of mitotic sister chromatid cohesion, telomeric(GO:0099403) mitotic sister chromatid cohesion, telomeric(GO:0099404) regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904907) negative regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904908) |
| 1.9 | 5.8 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 1.6 | 4.8 | GO:1901874 | negative regulation of post-translational protein modification(GO:1901874) |
| 1.6 | 11.0 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 1.5 | 10.4 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 1.2 | 6.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 1.2 | 3.7 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 1.2 | 3.5 | GO:1904387 | response to human chorionic gonadotropin(GO:0044752) cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 1.0 | 3.1 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 1.0 | 4.1 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 1.0 | 4.1 | GO:1904020 | negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of G-protein coupled receptor internalization(GO:1904020) negative regulation of G-protein coupled receptor internalization(GO:1904021) |
| 1.0 | 3.0 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) positive regulation of viral budding via host ESCRT complex(GO:1903774) regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 1.0 | 4.8 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.9 | 7.2 | GO:0090166 | regulation of Schwann cell differentiation(GO:0014038) Golgi disassembly(GO:0090166) |
| 0.9 | 4.4 | GO:2000542 | negative regulation of gastrulation(GO:2000542) |
| 0.8 | 3.1 | GO:0031179 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 0.8 | 3.8 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.7 | 4.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.7 | 6.1 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.7 | 5.3 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.6 | 1.9 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.6 | 1.3 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.6 | 9.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.6 | 1.8 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.6 | 2.4 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.6 | 8.3 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.6 | 1.7 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.5 | 3.7 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.5 | 2.5 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.5 | 3.0 | GO:1903899 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.5 | 1.8 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.4 | 3.0 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.4 | 1.7 | GO:0032072 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.4 | 1.3 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.4 | 3.7 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.4 | 8.1 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.4 | 1.2 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.4 | 7.6 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.4 | 5.1 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.4 | 3.4 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.4 | 2.5 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.3 | 4.9 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.3 | 0.7 | GO:0060003 | copper ion export(GO:0060003) |
| 0.3 | 0.7 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.3 | 14.2 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.3 | 0.9 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 0.3 | 1.5 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.3 | 2.6 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.3 | 2.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.3 | 13.4 | GO:0071451 | cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) |
| 0.3 | 2.1 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.3 | 0.8 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.3 | 0.8 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.3 | 7.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.3 | 1.8 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.3 | 3.6 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.2 | 0.7 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.2 | 3.4 | GO:1904871 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.2 | 1.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.2 | 5.8 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 2.4 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.2 | 2.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.2 | 0.6 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.2 | 5.2 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 2.0 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.2 | 2.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.2 | 1.1 | GO:0060356 | leucine import(GO:0060356) |
| 0.2 | 1.4 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.2 | 1.4 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.2 | 4.9 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.2 | 1.0 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.2 | 1.0 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.2 | 0.5 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.2 | 1.0 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.2 | 0.8 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.2 | 9.3 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.2 | 0.8 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.2 | 1.7 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 10.3 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 12.2 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.1 | 3.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 1.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 1.9 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 1.3 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 2.6 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.1 | 2.1 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 1.4 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.1 | 0.4 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 2.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 0.7 | GO:0046086 | AMP catabolic process(GO:0006196) adenosine biosynthetic process(GO:0046086) |
| 0.1 | 1.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 0.4 | GO:0061150 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) deltoid tuberosity development(GO:0035993) intermediate mesoderm development(GO:0048389) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) pattern specification involved in mesonephros development(GO:0061227) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in kidney development(GO:0072098) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.1 | 0.8 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 2.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 1.5 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.1 | 0.6 | GO:1900369 | regulation of RNA interference(GO:1900368) negative regulation of RNA interference(GO:1900369) |
| 0.1 | 1.0 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.3 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.1 | 4.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 3.5 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 3.8 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.1 | 0.5 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.1 | 3.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.5 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.1 | 0.8 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.1 | 4.3 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.1 | 0.8 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 2.1 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.1 | 0.5 | GO:0010748 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) plasma membrane long-chain fatty acid transport(GO:0015911) |
| 0.1 | 14.1 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.1 | 0.6 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 1.7 | GO:0016246 | RNA interference(GO:0016246) |
| 0.1 | 1.3 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.1 | 1.2 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.5 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 3.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 5.0 | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) |
| 0.1 | 1.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 1.6 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 1.0 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 1.4 | GO:0010510 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 2.5 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.1 | 5.4 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.1 | 0.7 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.2 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.1 | 1.8 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.1 | 0.6 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.1 | 0.4 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 3.9 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.1 | 0.7 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 | 0.6 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.1 | 1.7 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.1 | 3.2 | GO:0045661 | regulation of myoblast differentiation(GO:0045661) |
| 0.1 | 1.3 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 0.9 | GO:0002638 | negative regulation of immunoglobulin production(GO:0002638) |
| 0.1 | 3.8 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 0.1 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.1 | 3.9 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 2.3 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 | 0.6 | GO:1903671 | negative regulation of sprouting angiogenesis(GO:1903671) |
| 0.0 | 1.4 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.4 | GO:0034391 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 | 0.4 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.3 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:0048769 | skeletal muscle myosin thick filament assembly(GO:0030241) sarcomerogenesis(GO:0048769) |
| 0.0 | 0.1 | GO:1901318 | negative regulation of sperm motility(GO:1901318) |
| 0.0 | 0.3 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 2.1 | GO:0061053 | somite development(GO:0061053) |
| 0.0 | 0.3 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 2.4 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.6 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.6 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 1.5 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.6 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 3.1 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 0.5 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.2 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.3 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 1.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.5 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 1.4 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) |
| 0.0 | 1.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.5 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 1.1 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 1.4 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 2.9 | GO:0090002 | establishment of protein localization to plasma membrane(GO:0090002) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.3 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 2.3 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.0 | 14.7 | GO:0043066 | negative regulation of apoptotic process(GO:0043066) |
| 0.0 | 0.8 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 2.9 | GO:0007565 | female pregnancy(GO:0007565) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 1.2 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.8 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 1.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.5 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 1.4 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.9 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.2 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 1.8 | 7.2 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 1.6 | 11.0 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 1.5 | 13.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 1.5 | 4.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.2 | 3.5 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 1.1 | 6.6 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 1.1 | 5.3 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 1.1 | 3.2 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.9 | 3.5 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.8 | 3.4 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.8 | 8.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.8 | 9.0 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.6 | 6.0 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.6 | 3.0 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.5 | 3.8 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.5 | 2.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.5 | 3.7 | GO:0000243 | commitment complex(GO:0000243) |
| 0.5 | 5.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.4 | 7.4 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.4 | 1.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.4 | 1.5 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.3 | 2.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.3 | 7.8 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.3 | 3.0 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.3 | 2.6 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.3 | 4.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.3 | 3.4 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.2 | 1.4 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.2 | 4.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 2.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 7.1 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.2 | 0.8 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.2 | 0.8 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.2 | 1.0 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 4.8 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.2 | 1.3 | GO:0070761 | PCAF complex(GO:0000125) pre-snoRNP complex(GO:0070761) |
| 0.2 | 1.8 | GO:0044754 | autolysosome(GO:0044754) |
| 0.2 | 1.0 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 3.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 0.8 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.2 | 1.7 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 1.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 1.4 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 8.7 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 3.0 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 1.6 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 2.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 3.0 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.1 | 3.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 1.3 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 1.7 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 12.0 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 0.4 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 1.2 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 4.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 1.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 0.6 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 1.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 2.0 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 0.7 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 7.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 1.4 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 3.9 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.8 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 0.8 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 1.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 3.8 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 4.1 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 0.9 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.1 | 3.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.7 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 1.3 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 2.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.0 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 7.5 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 2.0 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 1.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 3.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 3.7 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 3.0 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 16.8 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 1.1 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 2.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 5.8 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 2.0 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.3 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 4.1 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 1.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 3.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 4.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 9.0 | GO:0030529 | intracellular ribonucleoprotein complex(GO:0030529) |
| 0.0 | 0.9 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 2.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 5.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.4 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.3 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 10.2 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 2.2 | 13.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 2.1 | 6.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 1.5 | 7.6 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 1.4 | 11.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 1.3 | 6.6 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 1.2 | 3.5 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 1.0 | 3.1 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) |
| 0.9 | 3.7 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.9 | 8.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.8 | 3.1 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.7 | 4.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.7 | 4.1 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.7 | 3.4 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.6 | 0.6 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.6 | 3.7 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.5 | 5.8 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.5 | 1.4 | GO:0000035 | acyl binding(GO:0000035) |
| 0.4 | 9.2 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.4 | 4.4 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.4 | 2.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.4 | 1.9 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.4 | 2.5 | GO:0102345 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.4 | 1.4 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.3 | 1.7 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.3 | 3.0 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.3 | 7.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.3 | 1.0 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.3 | 2.9 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.3 | 1.7 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.3 | 3.8 | GO:0046790 | virion binding(GO:0046790) |
| 0.3 | 0.8 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.3 | 10.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.3 | 1.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.2 | 1.7 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 1.7 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.2 | 0.7 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.2 | 1.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.2 | 1.3 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.2 | 2.0 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 2.6 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.2 | 3.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 3.0 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.2 | 0.6 | GO:0042806 | fucose binding(GO:0042806) |
| 0.2 | 3.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.2 | 0.6 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.2 | 0.6 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.2 | 12.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.2 | 6.0 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.2 | 3.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.2 | 1.0 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.2 | 4.9 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.2 | 1.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.2 | 1.0 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.2 | 4.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 0.8 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.2 | 8.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 1.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.2 | 7.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 0.6 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 1.7 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.7 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.8 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 2.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 1.3 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 2.6 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 4.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.2 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 6.3 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 0.3 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.1 | 0.8 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 4.5 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 6.1 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 1.6 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 5.0 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.9 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 6.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 4.9 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.4 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 8.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.5 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 3.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 1.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 3.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 1.5 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 1.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 1.7 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 1.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.6 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 1.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.8 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 2.4 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 0.2 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.1 | 1.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.3 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.1 | 0.4 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 1.9 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 1.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.2 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 0.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 3.2 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 3.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.6 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0004339 | glucan 1,4-alpha-glucosidase activity(GO:0004339) |
| 0.0 | 0.6 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 2.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.3 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.2 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.5 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.0 | 0.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 1.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.8 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.8 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.5 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 3.5 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 2.2 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 9.6 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.8 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.5 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 8.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.3 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0016899 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 0.0 | 7.3 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 5.6 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 1.0 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.1 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 2.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.5 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.8 | GO:0030165 | PDZ domain binding(GO:0030165) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 9.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 8.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 11.2 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.2 | 6.7 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.2 | 6.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 21.6 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.2 | 7.7 | PID ATM PATHWAY | ATM pathway |
| 0.2 | 13.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 5.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 6.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 5.1 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 1.0 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 4.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 8.5 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 1.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 4.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 0.9 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 6.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 8.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 0.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 1.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 3.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.5 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 1.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 2.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 3.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 2.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.3 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.5 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 14.9 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.8 | 9.0 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.4 | 3.0 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.4 | 6.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.4 | 13.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.4 | 26.5 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.3 | 10.8 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.3 | 5.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 0.5 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.2 | 4.1 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 3.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 5.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 2.9 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 5.9 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 7.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.2 | 4.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 2.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 3.8 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 3.0 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 8.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 5.0 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.1 | 1.7 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 11.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 9.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.2 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 2.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 3.0 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 3.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 3.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 17.6 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 1.6 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 2.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 4.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 0.4 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 1.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 0.8 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 0.9 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.0 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.8 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.7 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 1.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 1.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 2.1 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 2.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 2.0 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 1.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 6.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 4.8 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.6 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 4.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.5 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 4.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.7 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 1.5 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.0 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.0 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |