Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
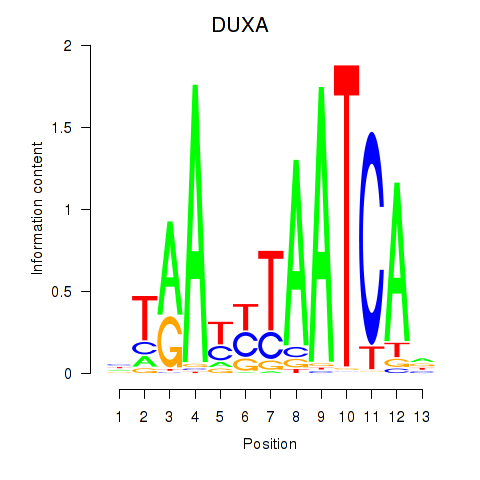
Results for DUXA
Z-value: 2.77
Transcription factors associated with DUXA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DUXA
|
ENSG00000258873.3 | DUXA |
Activity profile of DUXA motif
Sorted Z-values of DUXA motif
Network of associatons between targets according to the STRING database.
First level regulatory network of DUXA
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 11.0 | GO:0060221 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 1.6 | 4.8 | GO:1904899 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 1.4 | 5.5 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 1.4 | 4.1 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 1.3 | 5.2 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 1.3 | 3.9 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 1.2 | 7.4 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 1.2 | 3.5 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 1.0 | 3.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 1.0 | 3.1 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.8 | 2.5 | GO:0009720 | detection of hormone stimulus(GO:0009720) |
| 0.8 | 3.1 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.8 | 6.1 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.8 | 2.3 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.7 | 2.2 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.7 | 2.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.7 | 2.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.7 | 3.3 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.7 | 2.0 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.7 | 3.9 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.6 | 1.9 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.6 | 7.6 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.6 | 2.3 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.6 | 2.3 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.6 | 1.7 | GO:1902960 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.5 | 2.1 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.5 | 2.4 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.5 | 1.8 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.5 | 1.4 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.4 | 1.8 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.4 | 1.3 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.4 | 5.2 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.4 | 1.2 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.4 | 4.0 | GO:0006537 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.4 | 5.5 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.4 | 1.5 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.4 | 1.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.4 | 3.3 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.4 | 2.9 | GO:0006477 | protein sulfation(GO:0006477) heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.4 | 0.7 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.3 | 1.4 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.3 | 1.7 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.3 | 1.0 | GO:0071284 | copper ion export(GO:0060003) cellular response to lead ion(GO:0071284) regulation of electron carrier activity(GO:1904732) |
| 0.3 | 1.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.3 | 2.3 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.3 | 1.0 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.3 | 2.3 | GO:1902731 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.3 | 4.1 | GO:0070327 | lysine catabolic process(GO:0006554) thyroid hormone transport(GO:0070327) |
| 0.3 | 1.3 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.3 | 3.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.3 | 3.4 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.3 | 1.2 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.3 | 1.2 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.3 | 3.0 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.3 | 1.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.3 | 2.7 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.3 | 21.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.3 | 4.0 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.3 | 0.8 | GO:2000722 | mineralocorticoid receptor signaling pathway(GO:0031959) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) |
| 0.3 | 1.3 | GO:0010840 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.2 | 0.7 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.2 | 2.6 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.2 | 4.4 | GO:0015669 | gas transport(GO:0015669) oxygen transport(GO:0015671) |
| 0.2 | 0.7 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.2 | 0.4 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.2 | 1.3 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.2 | 0.7 | GO:0005988 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.2 | 0.7 | GO:0007343 | egg activation(GO:0007343) |
| 0.2 | 1.3 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.2 | 1.7 | GO:0015705 | iodide transport(GO:0015705) |
| 0.2 | 8.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.2 | 1.7 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.2 | 2.7 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.2 | 5.2 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.2 | 1.2 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.2 | 1.9 | GO:1902856 | negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.2 | 2.4 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.2 | 3.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.2 | 2.7 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.2 | 0.9 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.2 | 0.5 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.2 | 0.5 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.2 | 3.1 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.2 | 1.9 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.2 | 1.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.2 | 8.0 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.2 | 0.5 | GO:1903937 | response to acrylamide(GO:1903937) |
| 0.2 | 0.5 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.1 | 1.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 2.4 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) |
| 0.1 | 2.0 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 2.4 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.1 | 2.4 | GO:0071027 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.1 | 3.6 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.1 | 1.9 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 1.9 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 1.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.3 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.1 | 2.8 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 0.2 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.1 | 3.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 3.9 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 10.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 2.5 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.1 | 0.7 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 2.0 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 9.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 1.0 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.1 | 3.2 | GO:0061641 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 0.4 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.1 | 2.3 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.1 | 1.4 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 2.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 1.8 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 1.4 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 2.3 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.5 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.1 | 5.0 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 1.9 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.1 | 1.9 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.1 | 0.6 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 0.9 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 1.8 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 1.3 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 1.0 | GO:0010613 | positive regulation of cardiac muscle hypertrophy(GO:0010613) positive regulation of muscle hypertrophy(GO:0014742) |
| 0.1 | 1.0 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.1 | 0.5 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.1 | 0.5 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.1 | 0.6 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 1.0 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.1 | 1.8 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.1 | 0.8 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.1 | 1.1 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 2.5 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.1 | 17.7 | GO:0090305 | nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
| 0.1 | 4.5 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 40.9 | GO:0045596 | negative regulation of cell differentiation(GO:0045596) |
| 0.1 | 2.2 | GO:0006333 | chromatin assembly or disassembly(GO:0006333) |
| 0.0 | 1.1 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 1.2 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.4 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.9 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 | 1.4 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 1.4 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 1.1 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 1.5 | GO:0006378 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 2.6 | GO:0045814 | negative regulation of gene expression, epigenetic(GO:0045814) |
| 0.0 | 1.9 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 2.1 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 1.2 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.8 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.8 | GO:0010659 | cardiac muscle cell apoptotic process(GO:0010659) |
| 0.0 | 1.5 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.9 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 2.1 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.3 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 1.6 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.9 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 2.0 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 3.7 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 3.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.6 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.7 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.0 | 1.0 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.0 | 2.0 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 1.2 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.4 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 1.9 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.1 | GO:1904180 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.0 | 0.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.4 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.7 | 3.9 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.6 | 7.6 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.6 | 8.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.6 | 3.9 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.5 | 1.5 | GO:0031251 | PAN complex(GO:0031251) |
| 0.3 | 2.0 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.3 | 1.0 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.3 | 5.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.3 | 5.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.3 | 2.4 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.3 | 7.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.3 | 4.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.3 | 5.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.3 | 2.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 0.9 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.2 | 5.0 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.2 | 6.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 3.3 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.2 | 0.9 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.2 | 0.5 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) presynaptic cytoskeleton(GO:0099569) |
| 0.2 | 2.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 2.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 1.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 19.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 0.6 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.1 | 0.7 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 0.8 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 4.4 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 3.3 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.1 | 0.6 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 1.7 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.8 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 2.1 | GO:0043194 | node of Ranvier(GO:0033268) axon initial segment(GO:0043194) |
| 0.1 | 1.2 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.4 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 23.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 1.2 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.1 | 1.0 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 1.0 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 2.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.9 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 0.3 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 5.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 5.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.7 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 1.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.7 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 4.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.4 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 3.5 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 5.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.3 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 7.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 4.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 2.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.7 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 2.2 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 1.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 3.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.7 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 1.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 3.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.0 | GO:0034358 | protein-lipid complex(GO:0032994) plasma lipoprotein particle(GO:0034358) lipoprotein particle(GO:1990777) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 8.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.6 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 27.8 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.6 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 2.2 | GO:0000922 | spindle pole(GO:0000922) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 11.0 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 1.5 | 9.0 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 1.2 | 3.5 | GO:0004960 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 1.0 | 4.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 1.0 | 3.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 1.0 | 3.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.9 | 7.4 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.9 | 5.5 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.9 | 2.6 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.8 | 3.4 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.8 | 3.3 | GO:0004146 | dihydrofolate reductase activity(GO:0004146) |
| 0.8 | 8.0 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.8 | 3.1 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.7 | 3.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.7 | 2.9 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.7 | 2.1 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.7 | 2.0 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.7 | 2.7 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.6 | 4.4 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.6 | 1.9 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.6 | 2.5 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.6 | 2.4 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.6 | 2.3 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.5 | 2.4 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.5 | 1.8 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.5 | 2.7 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.5 | 5.0 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.4 | 1.3 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.4 | 3.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.4 | 1.5 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.4 | 3.9 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.3 | 1.0 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) phosphoglycerate kinase activity(GO:0004618) copper-transporting ATPase activity(GO:0043682) |
| 0.3 | 5.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.3 | 1.2 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.3 | 1.8 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.3 | 4.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.3 | 2.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.3 | 4.0 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.3 | 3.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.3 | 1.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.3 | 1.3 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.3 | 2.9 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.2 | 1.2 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.2 | 1.2 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.2 | 1.7 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.2 | 1.2 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.2 | 0.7 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.2 | 6.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 1.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 3.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 1.3 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.2 | 1.5 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.2 | 4.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.2 | 6.1 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.2 | 1.0 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.2 | 2.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 1.0 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.2 | 3.7 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 1.6 | GO:0030882 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.2 | 1.7 | GO:0005549 | odorant binding(GO:0005549) |
| 0.2 | 0.7 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.2 | 3.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 1.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 0.9 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.2 | 0.5 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.2 | 2.0 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.2 | 24.9 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.1 | 1.6 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.1 | 2.0 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 4.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.0 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 2.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 5.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 2.2 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 4.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 2.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 1.4 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 1.7 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 3.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 2.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 9.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 1.1 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.1 | 0.7 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 4.0 | GO:0008066 | glutamate receptor activity(GO:0008066) |
| 0.1 | 1.8 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 0.9 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 1.4 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 2.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 1.1 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 1.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.5 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.1 | 0.9 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 6.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.7 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 2.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 3.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.5 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 2.1 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 4.5 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.1 | 2.4 | GO:0005159 | insulin receptor binding(GO:0005158) insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 15.7 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 1.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 1.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.7 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.5 | GO:0042923 | opioid receptor activity(GO:0004985) neuropeptide binding(GO:0042923) |
| 0.0 | 2.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 7.8 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.6 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.9 | GO:0001164 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 1.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.8 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 1.0 | GO:0051184 | cofactor transporter activity(GO:0051184) |
| 0.0 | 0.8 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 1.0 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 5.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 5.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.3 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 1.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.5 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 26.1 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 2.5 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 4.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.3 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.1 | GO:0051378 | G-protein coupled serotonin receptor activity(GO:0004993) amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0070330 | aromatase activity(GO:0070330) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 11.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 7.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 17.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 1.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 3.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 3.5 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 5.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 1.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 4.2 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.1 | 4.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 3.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 3.5 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 1.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 2.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 6.5 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 4.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 4.1 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 3.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 2.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 6.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.9 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 1.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 2.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 3.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 2.1 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.9 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 6.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.6 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 8.0 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.3 | 5.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.3 | 3.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.3 | 3.9 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.3 | 6.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.3 | 7.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 9.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 2.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.2 | 4.0 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 4.1 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 6.2 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.2 | 1.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 4.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 1.8 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.2 | 1.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 4.8 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 2.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 11.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 1.9 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 3.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 3.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.0 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.1 | 3.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 2.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 0.9 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 1.6 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 1.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 4.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 2.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 1.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 2.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 2.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 4.0 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 1.8 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 1.1 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 2.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 5.5 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 2.2 | REACTOME PI3K EVENTS IN ERBB4 SIGNALING | Genes involved in PI3K events in ERBB4 signaling |
| 0.0 | 1.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 2.4 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.0 | 0.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.3 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 2.3 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 1.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 5.2 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 1.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |