Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
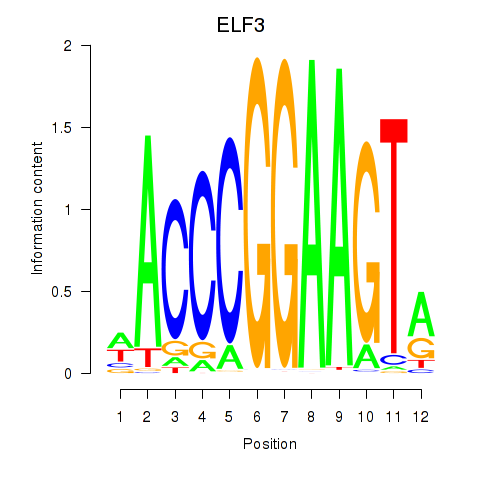
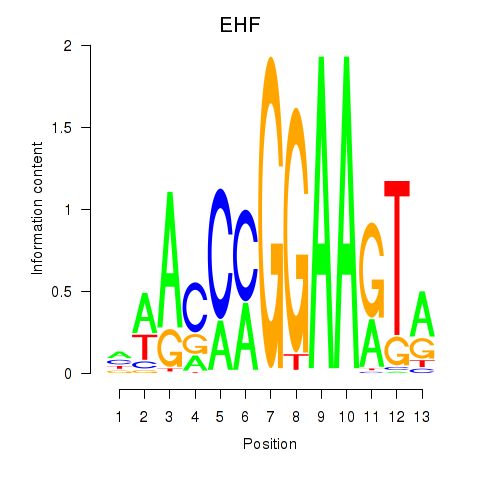
Results for ELF3_EHF
Z-value: 7.02
Transcription factors associated with ELF3_EHF
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ELF3
|
ENSG00000163435.16 | ELF3 |
|
EHF
|
ENSG00000135373.13 | EHF |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ELF3 | hg38_v1_chr1_+_202010575_202010593 | 0.28 | 3.4e-05 | Click! |
| EHF | hg38_v1_chr11_+_34621065_34621099, hg38_v1_chr11_+_34621109_34621135 | 0.11 | 1.2e-01 | Click! |
Activity profile of ELF3_EHF motif
Sorted Z-values of ELF3_EHF motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ELF3_EHF
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.5 | 50.1 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 7.3 | 21.9 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 5.3 | 16.0 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 5.3 | 5.3 | GO:0032056 | positive regulation of translation in response to stress(GO:0032056) positive regulation of translational initiation in response to stress(GO:0032058) |
| 5.1 | 15.4 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 4.6 | 18.4 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 4.5 | 13.6 | GO:0072716 | response to actinomycin D(GO:0072716) |
| 4.4 | 22.2 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 4.4 | 13.2 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 4.3 | 12.9 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 3.9 | 15.5 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 3.9 | 15.4 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 3.7 | 11.1 | GO:0061573 | endoplasmic reticulum polarization(GO:0061163) actin filament bundle retrograde transport(GO:0061573) actin filament bundle distribution(GO:0070650) |
| 3.5 | 31.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 3.3 | 16.3 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 3.2 | 9.6 | GO:0050720 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) interleukin-1 beta biosynthetic process(GO:0050720) response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) regulation of progesterone biosynthetic process(GO:2000182) |
| 3.0 | 15.2 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 3.0 | 45.1 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) |
| 2.9 | 8.7 | GO:2000832 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 2.8 | 22.4 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 2.8 | 8.4 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 2.7 | 8.2 | GO:0009996 | negative regulation of cell fate specification(GO:0009996) |
| 2.7 | 39.9 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 2.6 | 10.4 | GO:0019086 | late viral transcription(GO:0019086) |
| 2.5 | 7.6 | GO:0019075 | virus maturation(GO:0019075) |
| 2.5 | 10.0 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 2.4 | 9.8 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 2.4 | 12.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 2.4 | 7.1 | GO:0018282 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 2.3 | 7.0 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 2.3 | 9.0 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 2.3 | 6.8 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 2.2 | 15.7 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 2.2 | 13.0 | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 2.1 | 8.3 | GO:0071110 | protein biotinylation(GO:0009305) histone biotinylation(GO:0071110) |
| 1.9 | 5.8 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 1.9 | 5.8 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 1.9 | 24.8 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 1.9 | 9.5 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 1.9 | 36.0 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 1.9 | 20.4 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 1.8 | 10.9 | GO:1903232 | melanosome assembly(GO:1903232) |
| 1.8 | 5.4 | GO:0071988 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 1.7 | 3.5 | GO:0005988 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 1.7 | 8.6 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) negative regulation of vasoconstriction(GO:0045906) |
| 1.7 | 6.8 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 1.6 | 16.5 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 1.6 | 3.2 | GO:1904049 | regulation of spontaneous neurotransmitter secretion(GO:1904048) negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 1.6 | 4.8 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 1.6 | 4.7 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 1.5 | 4.6 | GO:1903538 | meiotic spindle elongation(GO:0051232) meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 1.5 | 9.1 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 1.5 | 7.6 | GO:2000870 | oocyte growth(GO:0001555) regulation of progesterone secretion(GO:2000870) |
| 1.5 | 7.4 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 1.5 | 4.4 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 1.5 | 13.2 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 1.4 | 4.3 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) glomerular visceral epithelial cell migration(GO:0090521) |
| 1.4 | 2.9 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 1.4 | 6.9 | GO:0010816 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 1.4 | 12.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 1.3 | 9.4 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 1.3 | 25.5 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 1.3 | 3.9 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 1.3 | 7.8 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 1.3 | 15.4 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 1.3 | 21.6 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 1.3 | 3.8 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 1.3 | 15.2 | GO:0097012 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 1.2 | 6.2 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 1.2 | 36.1 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 1.2 | 9.9 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 1.2 | 8.6 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 1.2 | 3.6 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 1.2 | 7.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 1.2 | 20.2 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 1.2 | 10.5 | GO:0006265 | DNA topological change(GO:0006265) |
| 1.1 | 10.3 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 1.1 | 30.5 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 1.1 | 3.4 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 1.1 | 4.5 | GO:2000845 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 1.1 | 4.5 | GO:0009233 | menaquinone metabolic process(GO:0009233) menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 1.1 | 4.5 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 1.1 | 7.7 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 1.1 | 12.1 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 1.1 | 6.6 | GO:0033504 | floor plate development(GO:0033504) |
| 1.0 | 3.1 | GO:0034091 | regulation of maintenance of sister chromatid cohesion(GO:0034091) regulation of maintenance of mitotic sister chromatid cohesion(GO:0034182) protein localization to site of double-strand break(GO:1990166) |
| 1.0 | 32.9 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 1.0 | 6.0 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 1.0 | 5.9 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 1.0 | 4.9 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 1.0 | 6.8 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 1.0 | 3.9 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.9 | 2.8 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.9 | 8.3 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.9 | 5.5 | GO:0001692 | histamine metabolic process(GO:0001692) histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.9 | 5.4 | GO:1904304 | regulation of gastro-intestinal system smooth muscle contraction(GO:1904304) positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.9 | 9.9 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.9 | 2.7 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.9 | 24.7 | GO:0050912 | detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.9 | 6.1 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.9 | 4.3 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.9 | 12.0 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.8 | 3.4 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.8 | 3.4 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.8 | 1.6 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.8 | 3.3 | GO:0010266 | response to vitamin B1(GO:0010266) |
| 0.8 | 6.3 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.8 | 2.3 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.8 | 6.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.8 | 15.2 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.8 | 3.8 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.8 | 8.3 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.7 | 5.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.7 | 35.6 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.7 | 3.7 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.7 | 5.1 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.7 | 6.6 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.7 | 2.2 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.7 | 16.5 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.7 | 2.1 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.7 | 5.7 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.7 | 2.1 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.7 | 2.1 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.7 | 7.7 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.7 | 2.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.7 | 2.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.7 | 2.7 | GO:0061358 | negative regulation of Wnt protein secretion(GO:0061358) |
| 0.7 | 6.8 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.7 | 8.2 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.7 | 1.4 | GO:0090182 | regulation of secretion of lysosomal enzymes(GO:0090182) |
| 0.7 | 2.7 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.7 | 8.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.7 | 4.0 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.7 | 5.9 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.7 | 19.7 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.7 | 2.0 | GO:0038109 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.7 | 13.7 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.6 | 2.6 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.6 | 5.8 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.6 | 5.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.6 | 1.9 | GO:1904640 | response to methionine(GO:1904640) |
| 0.6 | 8.2 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.6 | 6.8 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.6 | 3.7 | GO:0032252 | negative regulation of triglyceride catabolic process(GO:0010897) secretory granule localization(GO:0032252) |
| 0.6 | 1.9 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.6 | 32.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.6 | 4.7 | GO:0045836 | positive regulation of meiotic nuclear division(GO:0045836) |
| 0.6 | 12.2 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.6 | 8.7 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.6 | 4.0 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.6 | 12.3 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.6 | 1.7 | GO:2000097 | chronological cell aging(GO:0001300) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.6 | 3.9 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.6 | 9.9 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.6 | 7.2 | GO:0035878 | nail development(GO:0035878) |
| 0.6 | 2.2 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.5 | 7.7 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.5 | 2.2 | GO:0032641 | lymphotoxin A production(GO:0032641) positive regulation of mast cell cytokine production(GO:0032765) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.5 | 8.0 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.5 | 12.8 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.5 | 1.6 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.5 | 4.8 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.5 | 2.6 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.5 | 10.0 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.5 | 6.2 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.5 | 3.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.5 | 13.8 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.5 | 9.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.5 | 7.8 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.5 | 3.9 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.5 | 4.4 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.5 | 1.4 | GO:0060974 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) |
| 0.5 | 2.8 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.5 | 6.8 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.5 | 2.7 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.5 | 3.6 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.4 | 1.3 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.4 | 5.3 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.4 | 4.0 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.4 | 7.3 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.4 | 12.1 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.4 | 3.4 | GO:0015886 | heme transport(GO:0015886) |
| 0.4 | 13.4 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.4 | 6.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.4 | 2.8 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.4 | 5.3 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.4 | 7.2 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.4 | 16.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.4 | 5.6 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.4 | 4.0 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.4 | 3.1 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.4 | 10.5 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.4 | 4.6 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.4 | 4.2 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.4 | 2.3 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.4 | 4.9 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.4 | 4.9 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.4 | 3.4 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.4 | 4.5 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.4 | 6.7 | GO:0097152 | mesenchymal cell apoptotic process(GO:0097152) |
| 0.4 | 7.0 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.4 | 1.5 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.4 | 2.2 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.4 | 2.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.4 | 3.6 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.4 | 4.6 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.4 | 2.8 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.4 | 3.2 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.4 | 16.9 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.4 | 1.1 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.4 | 2.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.3 | 4.9 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.3 | 1.0 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.3 | 4.2 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.3 | 2.4 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.3 | 18.9 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.3 | 9.8 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.3 | 3.7 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.3 | 2.0 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.3 | 3.7 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.3 | 1.0 | GO:0039020 | pronephric nephron morphogenesis(GO:0039007) pronephric nephron tubule morphogenesis(GO:0039008) pronephric nephron tubule development(GO:0039020) pronephric duct development(GO:0039022) pronephric duct morphogenesis(GO:0039023) Kupffer's vesicle development(GO:0070121) |
| 0.3 | 2.3 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.3 | 2.2 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.3 | 6.4 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.3 | 2.9 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.3 | 0.9 | GO:0018874 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 0.3 | 3.4 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.3 | 5.3 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.3 | 9.1 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.3 | 1.5 | GO:1904354 | telomeric 3' overhang formation(GO:0031860) negative regulation of telomere capping(GO:1904354) |
| 0.3 | 1.2 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.3 | 2.1 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.3 | 1.5 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) |
| 0.3 | 2.0 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.3 | 13.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.3 | 1.1 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.3 | 1.4 | GO:0044791 | receptor-mediated endocytosis of virus by host cell(GO:0019065) modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.3 | 5.6 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.3 | 4.4 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.3 | 1.1 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.3 | 2.4 | GO:0050665 | hydrogen peroxide biosynthetic process(GO:0050665) |
| 0.3 | 1.3 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.3 | 3.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.3 | 5.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.3 | 2.3 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.3 | 1.0 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.3 | 1.8 | GO:1905123 | regulation of glucosylceramidase activity(GO:1905123) |
| 0.3 | 8.8 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.3 | 3.0 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.2 | 4.7 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.2 | 2.0 | GO:1900748 | positive regulation of cell migration by vascular endothelial growth factor signaling pathway(GO:0038089) positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.2 | 7.3 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.2 | 4.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 2.6 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.2 | 10.4 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.2 | 1.4 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.2 | 0.9 | GO:0010159 | specification of organ position(GO:0010159) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.2 | 28.1 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.2 | 0.7 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.2 | 6.6 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.2 | 9.8 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.2 | 1.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.2 | 3.5 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.2 | 21.7 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.2 | 3.4 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.2 | 4.7 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.2 | 9.7 | GO:0007602 | phototransduction(GO:0007602) |
| 0.2 | 9.5 | GO:0003091 | renal water homeostasis(GO:0003091) |
| 0.2 | 2.2 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.2 | 3.6 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.2 | 2.6 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.2 | 1.8 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 8.0 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.2 | 4.4 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.2 | 1.6 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.2 | 3.4 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.2 | 5.3 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.2 | 10.2 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.2 | 7.1 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.2 | 2.1 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.2 | 1.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.2 | 6.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.2 | 4.9 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.2 | 8.4 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.2 | 11.2 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.2 | 4.6 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.2 | 2.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.2 | 3.4 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.2 | 1.4 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.2 | 0.9 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.2 | 2.5 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.2 | 5.7 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.2 | 1.5 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.2 | 2.1 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.2 | 1.0 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.2 | 3.2 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.2 | 1.8 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.2 | 7.6 | GO:0009127 | purine nucleoside monophosphate biosynthetic process(GO:0009127) purine ribonucleoside monophosphate biosynthetic process(GO:0009168) |
| 0.2 | 2.5 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.2 | 10.8 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.2 | 1.3 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.2 | 5.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 1.2 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.2 | 0.8 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.2 | 2.3 | GO:1902221 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 3.5 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.1 | 4.8 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 5.3 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.1 | 0.9 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.1 | GO:1901628 | positive regulation of postsynaptic membrane organization(GO:1901628) regulation of receptor clustering(GO:1903909) positive regulation of receptor clustering(GO:1903911) |
| 0.1 | 2.9 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 4.7 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 1.7 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.1 | 3.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 1.9 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 | 3.8 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.1 | 0.7 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 1.5 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 | 0.7 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.1 | 0.8 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 2.4 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 2.1 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.1 | 1.7 | GO:0042269 | regulation of natural killer cell mediated cytotoxicity(GO:0042269) |
| 0.1 | 2.6 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 5.0 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.1 | 2.2 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.1 | 4.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.5 | GO:0002399 | MHC protein complex assembly(GO:0002396) MHC class II protein complex assembly(GO:0002399) |
| 0.1 | 0.7 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 4.7 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.1 | 8.5 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.1 | 3.1 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.1 | 1.1 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 1.4 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.1 | 1.1 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 0.9 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.4 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.1 | 6.7 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 0.7 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 0.5 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.1 | 4.9 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 | 6.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 3.1 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 1.0 | GO:0051969 | regulation of transmission of nerve impulse(GO:0051969) |
| 0.1 | 1.7 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.1 | 5.9 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 1.0 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.1 | 0.9 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 1.6 | GO:0051181 | cofactor transport(GO:0051181) |
| 0.1 | 0.6 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.1 | 3.8 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 2.5 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.1 | 10.2 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 1.0 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.1 | 11.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 2.8 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 1.2 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.1 | 0.4 | GO:0010616 | negative regulation of cardiac muscle adaptation(GO:0010616) regulation of lung blood pressure(GO:0014916) positive regulation of transforming growth factor beta3 production(GO:0032916) negative regulation of lung blood pressure(GO:0061767) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.1 | 0.2 | GO:0071442 | regulation of histone H3-K14 acetylation(GO:0071440) positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 | 2.8 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.1 | 1.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 0.9 | GO:0033081 | regulation of T cell differentiation in thymus(GO:0033081) regulation of thymocyte aggregation(GO:2000398) |
| 0.1 | 2.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 0.6 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) |
| 0.1 | 0.8 | GO:0038202 | TORC1 signaling(GO:0038202) |
| 0.1 | 0.5 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 0.7 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 0.2 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.1 | 0.9 | GO:0035411 | catenin import into nucleus(GO:0035411) |
| 0.1 | 0.5 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 3.4 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.2 | GO:0022615 | protein to membrane docking(GO:0022615) |
| 0.1 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 2.1 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.1 | 1.3 | GO:0042220 | response to cocaine(GO:0042220) |
| 0.1 | 0.4 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.1 | 0.5 | GO:0045910 | negative regulation of DNA recombination(GO:0045910) |
| 0.1 | 0.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0055099 | detection of hormone stimulus(GO:0009720) glycoprotein transport(GO:0034436) response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 1.5 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 4.8 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 1.1 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.1 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 | 1.3 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.7 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 4.1 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.0 | 0.7 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.0 | 1.3 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.2 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.5 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.2 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.2 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.7 | GO:0072378 | blood coagulation, intrinsic pathway(GO:0007597) blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.5 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 3.8 | GO:0007188 | adenylate cyclase-modulating G-protein coupled receptor signaling pathway(GO:0007188) |
| 0.0 | 0.1 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.0 | 3.8 | GO:0048705 | skeletal system morphogenesis(GO:0048705) |
| 0.0 | 0.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 2.0 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 2.5 | GO:0051384 | response to glucocorticoid(GO:0051384) |
| 0.0 | 0.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.8 | GO:0042472 | inner ear morphogenesis(GO:0042472) |
| 0.0 | 0.2 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 2.0 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 2.0 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.7 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 16.3 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 5.1 | 15.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 3.9 | 15.5 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 3.1 | 9.4 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 2.8 | 11.3 | GO:0000801 | central element(GO:0000801) |
| 2.8 | 44.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 2.6 | 15.7 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 2.4 | 12.1 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 2.3 | 9.1 | GO:1990879 | CST complex(GO:1990879) |
| 2.2 | 10.8 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 2.1 | 12.4 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 1.8 | 5.4 | GO:0055028 | cortical microtubule(GO:0055028) mitotic spindle astral microtubule(GO:0061673) |
| 1.6 | 6.3 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 1.6 | 20.5 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 1.4 | 4.3 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 1.4 | 6.9 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 1.4 | 1.4 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 1.2 | 3.7 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 1.2 | 5.9 | GO:0032449 | CBM complex(GO:0032449) |
| 1.2 | 4.8 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 1.2 | 14.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 1.2 | 5.8 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 1.2 | 8.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 1.2 | 5.8 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 1.1 | 4.4 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 1.0 | 3.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 1.0 | 4.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 1.0 | 4.1 | GO:0019034 | viral replication complex(GO:0019034) |
| 1.0 | 28.6 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.9 | 2.8 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.8 | 4.0 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.8 | 16.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.8 | 10.7 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.8 | 13.6 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.7 | 8.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.7 | 3.7 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.7 | 4.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.7 | 9.6 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.7 | 5.4 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.7 | 4.0 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.7 | 9.9 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.6 | 3.9 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.6 | 5.8 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.6 | 2.6 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.6 | 13.4 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.6 | 3.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.6 | 8.7 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.6 | 5.0 | GO:0070187 | telosome(GO:0070187) |
| 0.6 | 7.1 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.6 | 5.2 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.5 | 68.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.5 | 3.7 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.5 | 8.4 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.5 | 4.7 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.5 | 4.6 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.5 | 4.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.5 | 9.9 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.5 | 12.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.5 | 5.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.5 | 2.7 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.4 | 11.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.4 | 2.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.4 | 3.9 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.4 | 3.5 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.4 | 3.0 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.4 | 24.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.4 | 8.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.4 | 8.8 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.4 | 2.4 | GO:0060091 | kinocilium(GO:0060091) |
| 0.4 | 2.8 | GO:1990351 | transporter complex(GO:1990351) |
| 0.4 | 2.0 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.4 | 2.7 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.4 | 1.4 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.4 | 1.8 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.3 | 10.6 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.3 | 10.2 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.3 | 33.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.3 | 6.3 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.3 | 5.3 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.3 | 9.2 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.3 | 2.6 | GO:0034464 | BBSome(GO:0034464) |
| 0.3 | 7.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.3 | 6.1 | GO:0036038 | MKS complex(GO:0036038) |
| 0.3 | 4.7 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.3 | 0.9 | GO:0043259 | laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 0.3 | 5.9 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.3 | 48.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.3 | 2.7 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.3 | 17.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.3 | 6.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.3 | 4.8 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.2 | 6.1 | GO:0031095 | spectrin-associated cytoskeleton(GO:0014731) platelet dense tubular network membrane(GO:0031095) |
| 0.2 | 2.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 2.2 | GO:0044447 | axoneme part(GO:0044447) |
| 0.2 | 8.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.2 | 5.9 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 4.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.2 | 0.6 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.2 | 2.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 2.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 5.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 3.0 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.2 | 3.0 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.2 | 2.8 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 1.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.2 | 6.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 0.7 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.2 | 12.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.2 | 0.9 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.2 | 0.9 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.2 | 1.9 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 10.6 | GO:0043679 | axon terminus(GO:0043679) |
| 0.2 | 1.6 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.2 | 5.3 | GO:0005921 | gap junction(GO:0005921) |
| 0.2 | 11.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 1.0 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.2 | 1.5 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.2 | 2.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 15.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.2 | 7.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 1.7 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.9 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 8.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 4.1 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 30.0 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 1.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 2.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.6 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 12.4 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 2.7 | GO:0032589 | neuron projection membrane(GO:0032589) |
| 0.1 | 11.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 19.3 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 2.9 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 1.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 2.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 1.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 5.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 6.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 5.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 29.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 0.3 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 28.7 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 4.3 | GO:0031228 | intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 0.9 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 1.0 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 30.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 2.8 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 6.5 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 2.1 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.1 | 1.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.6 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 2.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.2 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 3.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 4.2 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 4.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 2.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 2.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 57.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 22.7 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.1 | 2.0 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 2.0 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.1 | 0.4 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 5.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 3.0 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 243.6 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.2 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.0 | 0.2 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 1.8 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 11.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.1 | GO:0097486 | late endosome lumen(GO:0031906) multivesicular body lumen(GO:0097486) |
| 0.0 | 0.7 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.7 | 48.7 | GO:0001129 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 5.3 | 16.0 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 3.9 | 15.4 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 3.5 | 10.5 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 3.4 | 13.8 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 3.3 | 10.0 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 3.3 | 10.0 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 3.2 | 19.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 3.2 | 19.1 | GO:0016807 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 2.9 | 8.7 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 2.8 | 8.4 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 2.7 | 8.0 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 2.6 | 28.7 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 2.6 | 10.3 | GO:0004146 | dihydrofolate reductase activity(GO:0004146) |
| 2.5 | 9.8 | GO:0004773 | steryl-sulfatase activity(GO:0004773) |
| 2.2 | 15.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 2.1 | 8.6 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 2.1 | 6.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 2.1 | 8.3 | GO:0004079 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 2.0 | 15.7 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 1.9 | 9.6 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 1.9 | 5.8 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 1.9 | 13.2 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 1.8 | 5.4 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 1.7 | 8.5 | GO:0004803 | transposase activity(GO:0004803) |
| 1.7 | 5.1 | GO:0052816 | medium-chain acyl-CoA hydrolase activity(GO:0052815) long-chain acyl-CoA hydrolase activity(GO:0052816) |
| 1.7 | 28.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 1.6 | 9.8 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 1.6 | 11.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 1.6 | 4.8 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 1.6 | 23.9 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 1.5 | 15.3 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 1.5 | 13.7 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 1.4 | 5.8 | GO:0032810 | sterol response element binding(GO:0032810) |
| 1.4 | 7.0 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 1.4 | 5.5 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) |
| 1.4 | 39.5 | GO:0071949 | FAD binding(GO:0071949) |
| 1.3 | 7.9 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 1.3 | 9.0 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 1.3 | 6.4 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 1.3 | 3.8 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 1.2 | 36.1 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 1.2 | 21.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 1.2 | 14.0 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 1.1 | 11.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 1.1 | 16.7 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 1.1 | 4.4 | GO:0052596 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 1.1 | 4.3 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 1.1 | 4.3 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 1.0 | 8.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 1.0 | 4.0 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 1.0 | 6.0 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 1.0 | 12.9 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 1.0 | 14.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.9 | 2.8 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.9 | 8.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.9 | 15.6 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.9 | 4.5 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.9 | 2.6 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.9 | 3.5 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.8 | 9.9 | GO:0008430 | selenium binding(GO:0008430) |
| 0.8 | 4.1 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.8 | 7.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.8 | 5.5 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.8 | 9.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.8 | 9.9 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.7 | 19.9 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.7 | 5.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.7 | 3.6 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.7 | 12.8 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.7 | 2.1 | GO:0046921 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.7 | 2.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.7 | 2.7 | GO:0004979 | beta-endorphin receptor activity(GO:0004979) morphine receptor activity(GO:0038047) |
| 0.7 | 3.4 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.7 | 6.8 | GO:0008061 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.7 | 8.7 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.7 | 6.0 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.6 | 4.5 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.6 | 3.8 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.6 | 8.6 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.6 | 6.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.6 | 22.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.6 | 8.4 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.6 | 20.2 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.6 | 32.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.6 | 10.0 | GO:0019864 | IgG binding(GO:0019864) |
| 0.6 | 13.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.6 | 2.3 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.6 | 5.8 | GO:0005549 | odorant binding(GO:0005549) |
| 0.6 | 11.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.6 | 2.3 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.5 | 4.4 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.5 | 1.6 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.5 | 9.6 | GO:0070513 | death domain binding(GO:0070513) |
| 0.5 | 4.8 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.5 | 1.6 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.5 | 2.6 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.5 | 5.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.5 | 3.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.5 | 3.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.5 | 4.7 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.5 | 2.5 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.5 | 15.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.5 | 5.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.5 | 3.4 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.5 | 9.8 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.5 | 6.8 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.5 | 6.8 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.5 | 7.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.5 | 2.3 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.5 | 2.3 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.5 | 10.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.5 | 12.7 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.4 | 2.2 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.4 | 28.6 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.4 | 4.2 | GO:0015386 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.4 | 2.5 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.4 | 2.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.4 | 29.1 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.4 | 3.7 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.4 | 6.1 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.4 | 6.5 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.4 | 5.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.4 | 6.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.4 | 2.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.4 | 3.5 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.4 | 7.3 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.4 | 10.8 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.4 | 6.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.4 | 6.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.4 | 3.7 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.4 | 2.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.4 | 1.4 | GO:0061714 | methotrexate binding(GO:0051870) folic acid receptor activity(GO:0061714) |
| 0.4 | 2.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.3 | 1.0 | GO:0001016 | RNA polymerase III regulatory region DNA binding(GO:0001016) |
| 0.3 | 1.0 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.3 | 1.7 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.3 | 6.2 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.3 | 1.0 | GO:0009032 | thymidine phosphorylase activity(GO:0009032) pyrimidine-nucleoside phosphorylase activity(GO:0016154) |
| 0.3 | 2.1 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.3 | 7.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.3 | 3.4 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.3 | 7.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 1.3 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.3 | 15.7 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.3 | 19.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.3 | 2.9 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.3 | 20.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.3 | 1.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.3 | 4.3 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.3 | 8.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.3 | 7.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.3 | 16.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.3 | 2.0 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.3 | 4.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.3 | 2.6 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.3 | 3.1 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.3 | 2.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.3 | 29.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.3 | 5.3 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.3 | 2.0 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.3 | 4.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.3 | 6.3 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.3 | 1.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.3 | 13.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.3 | 1.6 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.3 | 3.9 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.3 | 7.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.3 | 0.8 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.3 | 1.8 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 1.5 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.2 | 1.9 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.2 | 3.1 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.2 | 3.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 6.5 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.2 | 2.8 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.2 | 21.2 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.2 | 5.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 1.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 2.8 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.2 | 1.5 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.2 | 9.4 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.2 | 1.9 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.2 | 5.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 5.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 5.4 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.2 | 291.4 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.2 | 1.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.2 | 1.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 4.1 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 1.6 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.2 | 14.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 0.9 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.2 | 0.7 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.2 | 3.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 2.6 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 4.0 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.2 | 0.9 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.2 | 15.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.2 | 20.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.2 | 3.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 1.3 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.2 | 1.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.2 | 7.6 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.2 | 2.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 29.3 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 2.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 3.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 5.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 2.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 3.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 6.9 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.1 | 0.8 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 7.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 9.7 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 0.9 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 4.2 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 1.5 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 1.2 | GO:0031701 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 6.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 1.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 3.6 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 11.9 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.1 | 8.7 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 1.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 2.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 1.0 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 1.5 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 1.3 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 1.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 4.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 3.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 37.6 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 1.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.9 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 1.6 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 3.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 24.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 2.1 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 6.0 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 9.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 1.2 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.1 | 11.6 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.1 | 2.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 11.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 0.8 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 7.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 1.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.4 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 0.5 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 1.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 6.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.2 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) |
| 0.1 | 0.4 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 0.1 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.1 | 0.3 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.4 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 3.5 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 4.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.6 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 0.7 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.1 | 2.7 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.2 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.7 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 3.8 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 3.3 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 12.0 | GO:0046873 | metal ion transmembrane transporter activity(GO:0046873) |
| 0.0 | 1.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.2 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 1.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.1 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 1.1 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 26.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.7 | 12.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.6 | 40.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.5 | 17.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.5 | 3.8 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.5 | 16.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.4 | 23.5 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.4 | 14.9 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.4 | 18.6 | PID ATM PATHWAY | ATM pathway |
| 0.4 | 47.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.3 | 3.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.3 | 22.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.3 | 26.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.3 | 34.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.3 | 8.9 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.3 | 25.4 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.3 | 11.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 6.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.2 | 10.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 7.9 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.2 | 2.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 7.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.2 | 3.4 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.2 | 10.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.2 | 4.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 4.0 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.2 | 9.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.2 | 5.7 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 4.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 3.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 1.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 7.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 9.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 6.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 10.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 1.9 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 1.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 1.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 5.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 3.8 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 1.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 0.7 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 2.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 2.3 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 3.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 20.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 5.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 9.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 2.3 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.4 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.0 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.5 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.6 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 3.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 3.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID IL12 2PATHWAY | IL12-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 15.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.8 | 4.8 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.8 | 4.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.8 | 9.8 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.7 | 8.0 | REACTOME OPSINS | Genes involved in Opsins |
| 0.7 | 9.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.6 | 15.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.6 | 5.0 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.6 | 13.0 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.6 | 5.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.6 | 7.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.6 | 19.8 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.5 | 15.7 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.5 | 6.9 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.5 | 14.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.5 | 19.1 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.5 | 203.4 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.5 | 8.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.5 | 7.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.5 | 10.0 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.4 | 26.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.4 | 4.7 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.4 | 11.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.4 | 11.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.3 | 3.1 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.3 | 13.8 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.3 | 3.7 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.3 | 23.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.3 | 5.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.3 | 4.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.3 | 4.6 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.3 | 3.8 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.3 | 1.3 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.3 | 19.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.3 | 7.8 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.3 | 1.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.3 | 6.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 21.7 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 6.2 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.2 | 3.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 3.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 2.3 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.2 | 3.5 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.2 | 7.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.2 | 2.8 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 9.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 3.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 11.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 6.1 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.2 | 3.0 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 11.4 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.2 | 6.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.2 | 21.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 3.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 5.8 | REACTOME PI3K EVENTS IN ERBB2 SIGNALING | Genes involved in PI3K events in ERBB2 signaling |
| 0.1 | 2.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 5.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 3.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 3.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 2.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 4.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 1.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 0.8 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 3.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 4.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 4.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 10.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 4.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 13.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 3.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 2.4 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 0.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 2.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 2.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 3.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.7 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 6.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 14.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.3 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 3.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 1.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 8.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 0.7 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 2.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 3.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 3.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 2.1 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.0 | 0.2 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.2 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.3 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.9 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.9 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |