Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
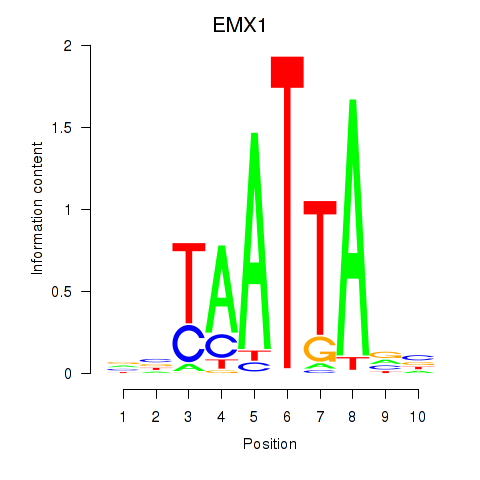
Results for EMX1
Z-value: 5.39
Transcription factors associated with EMX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EMX1
|
ENSG00000135638.14 | EMX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EMX1 | hg38_v1_chr2_+_72917489_72917525, hg38_v1_chr2_+_72916183_72916260 | 0.53 | 6.6e-17 | Click! |
Activity profile of EMX1 motif
Sorted Z-values of EMX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of EMX1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.0 | 24.0 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 5.7 | 17.1 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 4.8 | 19.2 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 4.5 | 57.9 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 3.5 | 10.4 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 3.4 | 58.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 3.4 | 10.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 3.3 | 13.4 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 2.8 | 8.4 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 2.6 | 7.7 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 2.5 | 25.1 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 2.4 | 14.4 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 2.4 | 16.6 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 2.4 | 7.1 | GO:0036166 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 2.2 | 6.7 | GO:0035425 | autocrine signaling(GO:0035425) |
| 2.1 | 6.4 | GO:0015847 | putrescine transport(GO:0015847) |
| 2.1 | 8.4 | GO:0031179 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 2.1 | 33.1 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 2.0 | 4.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 2.0 | 14.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 2.0 | 6.0 | GO:0015888 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 1.9 | 5.8 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 1.8 | 9.2 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 1.8 | 5.4 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 1.8 | 10.6 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 1.7 | 5.2 | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 1.7 | 10.4 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 1.7 | 11.7 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 1.6 | 8.2 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 1.6 | 4.9 | GO:0072675 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 1.6 | 6.4 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 1.6 | 4.8 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) sarcomerogenesis(GO:0048769) |
| 1.6 | 4.7 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 1.4 | 5.7 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 1.4 | 9.9 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 1.4 | 8.4 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 1.4 | 32.9 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 1.4 | 16.2 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.3 | 4.0 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 1.3 | 7.9 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 1.3 | 111.2 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 1.2 | 3.7 | GO:1900390 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 1.2 | 23.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 1.2 | 8.4 | GO:0007000 | nucleolus organization(GO:0007000) |
| 1.1 | 9.0 | GO:0061709 | reticulophagy(GO:0061709) |
| 1.1 | 7.9 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 1.1 | 3.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 1.1 | 21.7 | GO:0097186 | amelogenesis(GO:0097186) |
| 1.1 | 5.4 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 1.0 | 11.3 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 1.0 | 8.6 | GO:0030822 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) |
| 1.0 | 2.9 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.9 | 2.8 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.9 | 2.8 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.9 | 4.5 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.9 | 5.2 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.8 | 5.0 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.8 | 12.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.8 | 5.0 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.8 | 3.3 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.8 | 2.4 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.8 | 5.7 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.8 | 3.2 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.8 | 2.3 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.8 | 5.4 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.8 | 9.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.8 | 2.3 | GO:0033512 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.7 | 5.2 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.7 | 3.7 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.7 | 1.4 | GO:0070662 | mast cell proliferation(GO:0070662) |
| 0.7 | 8.5 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.7 | 5.6 | GO:0035878 | nail development(GO:0035878) |
| 0.7 | 3.5 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.7 | 7.7 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.7 | 2.0 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.7 | 5.3 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 0.7 | 2.0 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.7 | 5.9 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.6 | 1.9 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.6 | 3.7 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.6 | 9.9 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.6 | 8.8 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.6 | 2.9 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.6 | 1.7 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.6 | 3.4 | GO:0050955 | thermoception(GO:0050955) |
| 0.6 | 4.6 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.6 | 2.8 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.5 | 6.4 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.5 | 4.2 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.5 | 3.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.5 | 1.6 | GO:0019605 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 0.5 | 2.6 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.5 | 6.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.5 | 5.6 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.5 | 6.0 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.5 | 15.4 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.5 | 3.3 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.5 | 2.3 | GO:0070778 | L-aspartate transport(GO:0070778) L-aspartate transmembrane transport(GO:0089712) |
| 0.5 | 7.4 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.5 | 3.7 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.4 | 2.7 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.4 | 3.0 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.4 | 1.7 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.4 | 11.5 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.4 | 1.3 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.4 | 5.9 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.4 | 3.6 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.4 | 9.0 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.4 | 1.1 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.4 | 3.7 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.4 | 7.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.4 | 7.4 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.4 | 15.1 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.3 | 2.4 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.3 | 8.8 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.3 | 1.6 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.3 | 1.3 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) negative regulation of satellite cell differentiation(GO:1902725) |
| 0.3 | 2.8 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.3 | 4.9 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.3 | 4.2 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.3 | 14.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.3 | 1.5 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.3 | 3.8 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.3 | 4.6 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.3 | 28.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.3 | 4.6 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.2 | 6.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 22.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.2 | 0.9 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.2 | 6.6 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.2 | 3.6 | GO:0043691 | reverse cholesterol transport(GO:0043691) |
| 0.2 | 3.8 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.2 | 7.5 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.2 | 1.5 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.2 | 0.4 | GO:0060685 | regulation of prostatic bud formation(GO:0060685) negative regulation of prostatic bud formation(GO:0060686) |
| 0.2 | 34.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 1.3 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.2 | 2.4 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.2 | 0.6 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.2 | 4.9 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.2 | 0.6 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
| 0.2 | 8.2 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.2 | 2.0 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.2 | 2.1 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.2 | 1.2 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.2 | 3.8 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.2 | 7.6 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.2 | 4.4 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.2 | 4.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.2 | 10.7 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.2 | 1.3 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.2 | 5.4 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.2 | 2.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.2 | 0.5 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.2 | 2.0 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 3.2 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 5.4 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) regulation of adenylate cyclase activity(GO:0045761) |
| 0.1 | 0.7 | GO:0046618 | drug export(GO:0046618) |
| 0.1 | 6.0 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 1.8 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 4.8 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 3.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 5.0 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.1 | 4.7 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.1 | 0.4 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.1 | 7.5 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 0.9 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 3.2 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 2.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 6.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 1.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 1.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.3 | GO:1903438 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.1 | 1.3 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 2.6 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 0.2 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 0.7 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 2.6 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.1 | 3.8 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 3.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 3.4 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 2.4 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.1 | 3.5 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 0.6 | GO:0009048 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.1 | 1.1 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 7.1 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.1 | 2.0 | GO:0003091 | renal water homeostasis(GO:0003091) |
| 0.1 | 2.7 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 9.7 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.1 | 4.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 3.8 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.1 | 5.1 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.1 | 0.7 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 5.1 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 2.4 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 0.4 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.1 | 4.2 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 4.1 | GO:0048477 | oogenesis(GO:0048477) |
| 0.1 | 8.4 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.1 | 1.7 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 | 4.0 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 2.3 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 1.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 10.3 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 1.0 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.7 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.9 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.9 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.6 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 4.6 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 4.4 | GO:2000117 | negative regulation of cysteine-type endopeptidase activity(GO:2000117) |
| 0.0 | 25.3 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.6 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.2 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 3.4 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 3.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 1.3 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 | 1.2 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 1.1 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 2.5 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 1.0 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 57.9 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 3.8 | 11.5 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 3.6 | 58.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 3.5 | 10.4 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 2.1 | 6.2 | GO:0043159 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) acrosomal matrix(GO:0043159) |
| 1.9 | 11.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 1.9 | 5.6 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 1.4 | 20.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 1.4 | 21.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 1.3 | 7.9 | GO:0033063 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 1.3 | 10.4 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 1.3 | 7.6 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 1.2 | 14.4 | GO:0016013 | syntrophin complex(GO:0016013) |
| 1.1 | 5.4 | GO:0014802 | terminal cisterna(GO:0014802) |
| 1.0 | 5.9 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.9 | 2.6 | GO:0036027 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.8 | 2.3 | GO:0098831 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) presynaptic cytoskeleton(GO:0099569) |
| 0.8 | 6.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.8 | 3.0 | GO:0071546 | pi-body(GO:0071546) |
| 0.7 | 14.4 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.7 | 4.2 | GO:0002177 | manchette(GO:0002177) |
| 0.7 | 4.9 | GO:0060091 | kinocilium(GO:0060091) |
| 0.6 | 5.0 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.6 | 7.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.6 | 7.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.6 | 33.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.6 | 11.7 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.5 | 26.4 | GO:0031430 | M band(GO:0031430) |
| 0.5 | 12.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.4 | 67.2 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.4 | 2.7 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.4 | 6.1 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.4 | 3.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.4 | 6.0 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.4 | 2.4 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.4 | 9.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.4 | 15.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.4 | 20.6 | GO:0002102 | podosome(GO:0002102) |
| 0.4 | 7.7 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.4 | 4.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.3 | 2.4 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.3 | 5.9 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.3 | 5.8 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.3 | 9.0 | GO:0031672 | A band(GO:0031672) |
| 0.3 | 2.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.3 | 21.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.3 | 5.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.3 | 5.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.2 | 1.5 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.2 | 5.7 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.2 | 5.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.2 | 3.6 | GO:0016342 | fascia adherens(GO:0005916) catenin complex(GO:0016342) |
| 0.2 | 3.7 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.2 | 4.0 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.2 | 6.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.2 | 18.1 | GO:0042641 | actomyosin(GO:0042641) |
| 0.2 | 32.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.2 | 43.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.2 | 5.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 3.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 8.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 6.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 1.7 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.1 | 5.8 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 4.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 3.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 2.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 16.5 | GO:0045121 | membrane raft(GO:0045121) |
| 0.1 | 0.9 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 9.3 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 1.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 1.7 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 10.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 3.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 4.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 7.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 2.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 4.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 0.6 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 2.5 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 9.3 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 144.4 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 93.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 9.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 1.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 4.5 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.1 | 9.1 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 0.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 9.8 | GO:0098793 | presynapse(GO:0098793) |
| 0.1 | 3.4 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 3.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 4.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 2.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 12.8 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.3 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 5.0 | GO:0031966 | mitochondrial membrane(GO:0031966) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 9.1 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 3.0 | 18.0 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 2.7 | 10.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 2.6 | 10.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 2.5 | 7.6 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 2.5 | 7.6 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 2.3 | 11.3 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 2.2 | 66.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 2.1 | 6.4 | GO:0015203 | polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) |
| 2.1 | 8.4 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 2.0 | 6.0 | GO:0015403 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) |
| 2.0 | 24.0 | GO:0008430 | selenium binding(GO:0008430) |
| 1.9 | 5.7 | GO:1990254 | keratin filament binding(GO:1990254) |
| 1.8 | 9.0 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 1.7 | 114.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 1.4 | 16.6 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 1.4 | 13.7 | GO:0004064 | arylesterase activity(GO:0004064) |
| 1.4 | 21.8 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 1.3 | 4.0 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 1.3 | 7.7 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 1.2 | 39.9 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 1.2 | 8.5 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 1.2 | 21.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 1.2 | 7.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 1.2 | 6.0 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 1.2 | 16.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 1.2 | 11.5 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 1.1 | 4.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 1.1 | 6.4 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 1.1 | 5.3 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 1.1 | 3.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 1.0 | 4.0 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 1.0 | 12.5 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.9 | 21.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.9 | 5.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.9 | 11.7 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.9 | 2.7 | GO:0001034 | RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.9 | 8.9 | GO:0005549 | odorant binding(GO:0005549) |
| 0.9 | 7.7 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.8 | 5.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.8 | 7.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.8 | 19.2 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.8 | 6.9 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.7 | 9.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.7 | 2.0 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.7 | 9.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.6 | 1.9 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.6 | 8.6 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.6 | 12.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.6 | 2.4 | GO:0004773 | steryl-sulfatase activity(GO:0004773) |
| 0.6 | 48.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.6 | 3.5 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.6 | 1.7 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.5 | 4.9 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.5 | 5.4 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.5 | 7.9 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.5 | 6.9 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.5 | 1.4 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.5 | 1.9 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.5 | 9.9 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.5 | 7.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.5 | 2.3 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.5 | 5.5 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.4 | 26.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.4 | 9.9 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.4 | 5.6 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.4 | 8.8 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.4 | 8.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.4 | 38.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.4 | 1.1 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.4 | 10.9 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.4 | 10.2 | GO:0071949 | FAD binding(GO:0071949) |
| 0.3 | 2.8 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.3 | 31.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.3 | 1.7 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.3 | 3.7 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.3 | 2.0 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.3 | 9.2 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.3 | 2.8 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.3 | 1.5 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.3 | 1.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.3 | 3.8 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.3 | 6.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.3 | 4.9 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.3 | 2.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.3 | 2.9 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.3 | 5.0 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.3 | 9.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.3 | 55.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.3 | 7.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.2 | 3.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.2 | 2.8 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.2 | 38.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.2 | 6.6 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.2 | 19.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.2 | 3.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 1.2 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.2 | 1.8 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.2 | 33.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.2 | 0.6 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.2 | 1.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 2.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.2 | 1.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) protein histidine kinase activity(GO:0004673) |
| 0.2 | 17.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.2 | 1.4 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.2 | 8.8 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.2 | 7.1 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.2 | 1.6 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.2 | 5.9 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.2 | 3.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 1.7 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 2.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 1.3 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 1.3 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 1.6 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 18.0 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.1 | 3.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 11.1 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 2.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 12.6 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 8.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 1.4 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 10.6 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.1 | 32.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 4.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 5.7 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 2.3 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 1.5 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.1 | 2.6 | GO:0042166 | acetylcholine-gated cation channel activity(GO:0022848) acetylcholine binding(GO:0042166) |
| 0.1 | 1.7 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 5.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 8.9 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 1.7 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 15.2 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 2.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 4.9 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 1.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.9 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 0.6 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.5 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.6 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 3.3 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 3.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 9.9 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 7.6 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.1 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.0 | 0.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 18.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.2 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 61.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.5 | 10.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.4 | 33.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.3 | 22.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.3 | 8.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.3 | 12.5 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.3 | 18.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.3 | 8.6 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.3 | 5.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.3 | 3.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 13.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 7.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 5.4 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.2 | 23.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.2 | 2.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.2 | 14.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.2 | 9.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 30.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 7.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 8.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 8.5 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 34.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 5.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 7.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 12.2 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 35.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 3.2 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 5.0 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 0.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 0.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 1.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 3.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 2.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 2.4 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.9 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 57.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 2.1 | 49.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.9 | 13.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.8 | 16.8 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.7 | 21.8 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.6 | 10.4 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.5 | 5.9 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.5 | 6.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.5 | 38.8 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.5 | 12.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.4 | 14.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.4 | 16.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.4 | 19.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.3 | 13.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.3 | 7.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.3 | 5.6 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.3 | 19.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.3 | 11.7 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.3 | 23.0 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.3 | 10.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 7.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 7.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 7.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.2 | 8.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 5.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.2 | 3.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.2 | 3.2 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.2 | 5.6 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.2 | 5.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 4.0 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.2 | 2.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 4.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.2 | 8.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 1.4 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.2 | 7.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 4.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 4.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 4.7 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.2 | 13.0 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.2 | 33.0 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.2 | 3.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 2.0 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.2 | 8.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 6.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 4.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 6.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 19.4 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.1 | 6.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.6 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 2.6 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 0.6 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 4.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.1 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 6.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 5.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 3.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 6.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 6.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 1.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 3.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 7.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 3.4 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 2.6 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 5.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.7 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 1.7 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 1.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.0 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |