Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
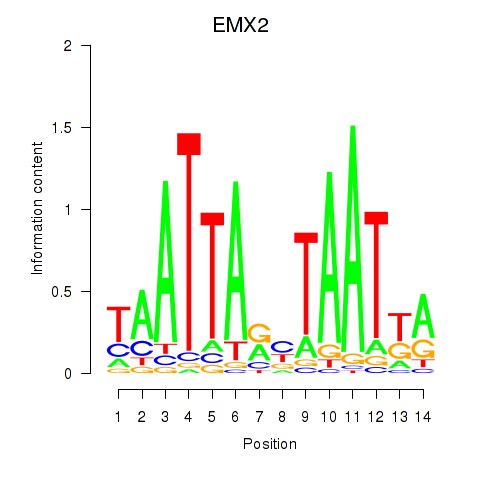
Results for EMX2
Z-value: 5.24
Transcription factors associated with EMX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EMX2
|
ENSG00000170370.12 | EMX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EMX2 | hg38_v1_chr10_+_117543567_117543658, hg38_v1_chr10_+_117542721_117542754, hg38_v1_chr10_+_117542416_117542445 | 0.23 | 4.8e-04 | Click! |
Activity profile of EMX2 motif
Sorted Z-values of EMX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of EMX2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.4 | 46.2 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 3.2 | 22.5 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 3.1 | 9.3 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 2.4 | 7.3 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 2.3 | 7.0 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 2.1 | 14.4 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 2.0 | 12.3 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 2.0 | 25.9 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 1.8 | 7.3 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 1.7 | 13.8 | GO:0014055 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) |
| 1.7 | 6.8 | GO:0050758 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) cellular response to folic acid(GO:0071231) |
| 1.7 | 8.5 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 1.7 | 16.8 | GO:0009449 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 1.6 | 8.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 1.4 | 6.9 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 1.4 | 21.7 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 1.2 | 7.4 | GO:0097338 | response to clozapine(GO:0097338) |
| 1.2 | 13.4 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 1.2 | 6.0 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 1.2 | 6.0 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 1.2 | 5.9 | GO:0071727 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 1.1 | 3.4 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 1.0 | 3.1 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 1.0 | 2.9 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 1.0 | 6.8 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.9 | 5.6 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.9 | 3.7 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.9 | 3.7 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.9 | 2.8 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.8 | 2.5 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.8 | 5.7 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.8 | 6.1 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.8 | 3.0 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.7 | 5.2 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.7 | 2.8 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.7 | 8.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.7 | 18.0 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.7 | 16.5 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.6 | 1.9 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.6 | 4.5 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.6 | 7.8 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.6 | 4.0 | GO:1902731 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.5 | 5.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.5 | 9.4 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.5 | 2.5 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.5 | 4.3 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.5 | 6.8 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.5 | 1.4 | GO:0032761 | negative regulation of T cell tolerance induction(GO:0002665) negative regulation of T cell anergy(GO:0002668) negative regulation of lymphocyte anergy(GO:0002912) regulation of lymphotoxin A production(GO:0032681) positive regulation of lymphotoxin A production(GO:0032761) regulation of lymphotoxin A biosynthetic process(GO:0043016) positive regulation of lymphotoxin A biosynthetic process(GO:0043017) |
| 0.5 | 17.8 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.5 | 2.7 | GO:0050955 | thermoception(GO:0050955) |
| 0.5 | 10.4 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.4 | 3.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.4 | 18.9 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.4 | 2.0 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.4 | 1.2 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.4 | 4.2 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.4 | 1.9 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.4 | 2.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.4 | 4.5 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.4 | 8.7 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.4 | 4.5 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.4 | 1.1 | GO:1902161 | transepithelial water transport(GO:0035377) positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.4 | 1.4 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.3 | 10.5 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.3 | 3.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.3 | 25.6 | GO:0007032 | endosome organization(GO:0007032) |
| 0.3 | 8.3 | GO:0006067 | ethanol metabolic process(GO:0006067) |
| 0.3 | 4.9 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.3 | 7.3 | GO:0006833 | water transport(GO:0006833) |
| 0.3 | 3.8 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.3 | 1.0 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.3 | 0.8 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.3 | 6.5 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) |
| 0.3 | 8.8 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.3 | 0.8 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.2 | 0.7 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) regulation of locomotion involved in locomotory behavior(GO:0090325) |
| 0.2 | 2.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.2 | 6.9 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.2 | 3.8 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.2 | 20.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.2 | 12.3 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.2 | 2.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 2.3 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.2 | 1.5 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.2 | 7.0 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.2 | 2.4 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 3.2 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.2 | 0.8 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.2 | 1.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.2 | 8.1 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.2 | 1.4 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.2 | 0.9 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.2 | 13.4 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.2 | 9.3 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.1 | 4.9 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 3.6 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 2.2 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 2.0 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.1 | 7.9 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 0.9 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.1 | 0.3 | GO:1902617 | response to fluoride(GO:1902617) |
| 0.1 | 1.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 1.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 1.1 | GO:0045794 | negative regulation of cell volume(GO:0045794) smooth muscle contraction involved in micturition(GO:0060083) |
| 0.1 | 6.2 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 11.7 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 0.1 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.1 | 1.7 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 7.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 1.9 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 1.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 2.4 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.1 | 1.7 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 3.6 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 2.0 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 1.1 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.1 | 0.6 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 | 1.6 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.1 | 10.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.5 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 1.9 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.1 | 5.3 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.1 | 2.9 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 1.2 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 4.8 | GO:0071357 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 4.3 | GO:0098869 | cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.9 | GO:0030825 | positive regulation of cGMP metabolic process(GO:0030825) positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 2.1 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.0 | 1.5 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.4 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 1.1 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 3.1 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 1.8 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.3 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.2 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 1.9 | GO:0002763 | positive regulation of myeloid leukocyte differentiation(GO:0002763) |
| 0.0 | 0.1 | GO:2000523 | defense response to nematode(GO:0002215) regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.9 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 1.4 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.4 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 1.6 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 1.0 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 1.6 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.1 | GO:1990822 | basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.9 | GO:0007588 | excretion(GO:0007588) |
| 0.0 | 2.7 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.1 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 1.2 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.0 | 0.7 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.0 | 48.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 1.4 | 12.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 1.3 | 14.4 | GO:0043203 | axon hillock(GO:0043203) |
| 1.0 | 5.7 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.8 | 3.4 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.8 | 2.5 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.8 | 5.6 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.7 | 2.0 | GO:0036028 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.7 | 4.0 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.7 | 5.9 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.6 | 7.8 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.6 | 6.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.6 | 7.7 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.5 | 8.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.5 | 17.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.5 | 11.2 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.5 | 6.0 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.4 | 18.0 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.4 | 7.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.4 | 21.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.4 | 1.1 | GO:0071752 | dimeric IgA immunoglobulin complex(GO:0071750) secretory dimeric IgA immunoglobulin complex(GO:0071752) IgM immunoglobulin complex(GO:0071753) IgM immunoglobulin complex, circulating(GO:0071754) pentameric IgM immunoglobulin complex(GO:0071756) |
| 0.4 | 2.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.3 | 4.0 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.3 | 8.2 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 5.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.2 | 1.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.2 | 2.9 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 5.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 1.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 3.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 3.8 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 2.9 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.1 | 2.5 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 3.4 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 2.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 3.1 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 2.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 1.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 0.5 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 3.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 5.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 18.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 4.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.8 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 9.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 9.1 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 8.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 3.8 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 8.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 2.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 3.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 2.7 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 7.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 7.1 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 1.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.3 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 2.8 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.9 | 48.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 4.5 | 26.9 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 2.7 | 8.2 | GO:0016497 | substance K receptor activity(GO:0016497) |
| 2.1 | 8.5 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 1.8 | 7.3 | GO:0004031 | aldehyde oxidase activity(GO:0004031) ethanol binding(GO:0035276) |
| 1.8 | 5.3 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 1.8 | 7.0 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 1.7 | 6.9 | GO:0004802 | transketolase activity(GO:0004802) |
| 1.7 | 6.8 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 1.7 | 5.0 | GO:0052854 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 1.5 | 5.9 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 1.4 | 5.6 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 1.3 | 3.9 | GO:0042806 | fucose binding(GO:0042806) |
| 1.2 | 13.4 | GO:0004568 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 1.2 | 16.8 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 1.1 | 18.0 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 1.1 | 3.4 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 1.0 | 3.0 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 1.0 | 5.9 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.9 | 8.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.8 | 4.2 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.8 | 3.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.7 | 5.2 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.7 | 2.2 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.7 | 2.7 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.6 | 3.6 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.6 | 7.0 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.5 | 26.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.5 | 3.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.5 | 3.8 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.5 | 2.3 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.5 | 3.7 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.5 | 2.3 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.5 | 7.4 | GO:0004691 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.5 | 7.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.4 | 3.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.4 | 1.3 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.4 | 7.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.4 | 4.4 | GO:0005549 | odorant binding(GO:0005549) |
| 0.4 | 6.5 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.4 | 10.4 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.4 | 9.3 | GO:0051430 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.4 | 1.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.3 | 4.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.3 | 1.9 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.3 | 1.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.3 | 17.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.3 | 0.9 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.3 | 5.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.3 | 4.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.3 | 3.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.3 | 5.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.2 | 2.4 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.2 | 15.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.2 | 3.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.2 | 1.6 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 3.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 2.0 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.2 | 3.8 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 1.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.2 | 40.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.2 | 4.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 3.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.2 | 0.9 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.2 | 15.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.2 | 1.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 31.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.2 | 3.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 4.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 14.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 4.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 2.6 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 10.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 6.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 1.4 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 0.7 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 1.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 3.7 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 1.4 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 1.6 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 1.0 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.1 | 1.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 2.4 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 3.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.6 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 5.4 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 2.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 8.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 1.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 8.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 1.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 1.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.8 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 40.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 2.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 3.5 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 2.7 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.2 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 3.5 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.9 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 6.2 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 3.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.4 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.4 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 4.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.7 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 4.3 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 1.6 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 11.8 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 1.8 | GO:0016782 | transferase activity, transferring sulfur-containing groups(GO:0016782) |
| 0.0 | 0.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 1.6 | GO:0022843 | voltage-gated cation channel activity(GO:0022843) |
| 0.0 | 4.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 4.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.9 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 3.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 10.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 4.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 7.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 6.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 9.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 7.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 9.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 2.3 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 13.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 2.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 6.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 23.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 1.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 2.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.4 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.9 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 2.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 3.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 26.9 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 1.0 | 9.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.8 | 18.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.6 | 7.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.5 | 17.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.5 | 18.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.5 | 5.9 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.4 | 8.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.3 | 11.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.3 | 6.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.3 | 47.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.3 | 7.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.3 | 22.0 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.3 | 5.6 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.3 | 5.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 3.7 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 3.0 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.2 | 13.5 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.2 | 15.7 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 3.6 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.2 | 3.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 3.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 2.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 3.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 4.2 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 4.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 8.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 2.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 4.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 2.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.9 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 4.0 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 12.6 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 1.4 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 3.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.9 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 3.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 8.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 5.7 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 1.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 7.4 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |