Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for EN2_GBX2_LBX2
Z-value: 2.48
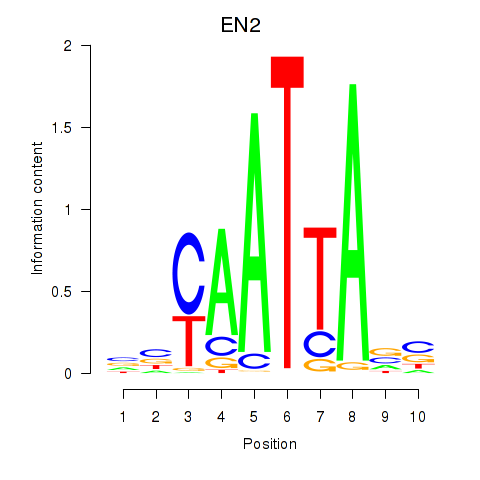
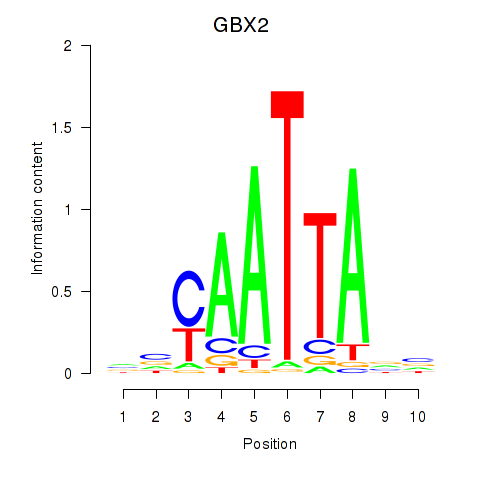
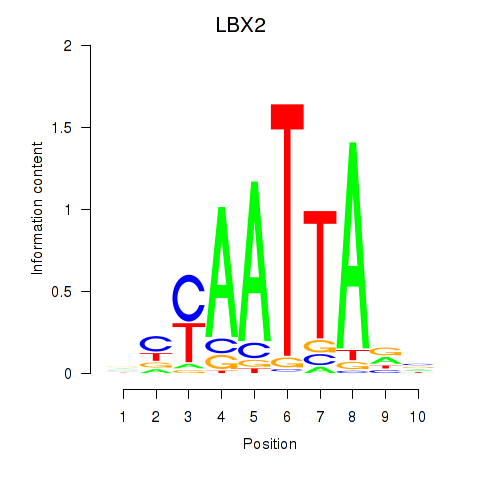
Transcription factors associated with EN2_GBX2_LBX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EN2
|
ENSG00000164778.4 | EN2 |
|
GBX2
|
ENSG00000168505.7 | GBX2 |
|
LBX2
|
ENSG00000179528.16 | LBX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EN2 | hg38_v1_chr7_+_155458129_155458129 | 0.28 | 3.0e-05 | Click! |
| GBX2 | hg38_v1_chr2_-_236168376_236168395 | 0.20 | 3.7e-03 | Click! |
Activity profile of EN2_GBX2_LBX2 motif
Sorted Z-values of EN2_GBX2_LBX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of EN2_GBX2_LBX2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.3 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 2.7 | 26.6 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 2.5 | 7.4 | GO:1904395 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 1.5 | 4.6 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 1.5 | 13.1 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 1.2 | 3.7 | GO:0071140 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 1.2 | 11.0 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 1.0 | 2.9 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 1.0 | 3.8 | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.9 | 12.7 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.7 | 4.7 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.6 | 6.5 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.6 | 1.8 | GO:0034183 | negative regulation of maintenance of sister chromatid cohesion(GO:0034092) negative regulation of maintenance of mitotic sister chromatid cohesion(GO:0034183) protein auto-ADP-ribosylation(GO:0070213) maintenance of mitotic sister chromatid cohesion, telomeric(GO:0099403) mitotic sister chromatid cohesion, telomeric(GO:0099404) regulation of telomeric DNA binding(GO:1904742) regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904907) negative regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904908) |
| 0.4 | 9.8 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.4 | 2.7 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.4 | 1.3 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.4 | 2.0 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.4 | 3.8 | GO:0021814 | cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) |
| 0.3 | 1.0 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.3 | 8.6 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.3 | 1.5 | GO:0061743 | motor learning(GO:0061743) |
| 0.2 | 1.0 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.2 | 14.4 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.2 | 1.6 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.2 | 1.3 | GO:1902231 | positive regulation of keratinocyte apoptotic process(GO:1902174) positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.2 | 15.5 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 1.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 1.8 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 2.5 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.7 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.1 | 8.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 6.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 2.9 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 0.2 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 2.1 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.3 | GO:0030497 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 2.5 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.1 | 0.2 | GO:0072366 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.1 | 9.5 | GO:0006757 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.1 | 7.2 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.1 | 1.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 2.2 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.6 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 2.0 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.8 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 1.9 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.0 | 0.2 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 5.5 | GO:0007422 | peripheral nervous system development(GO:0007422) |
| 0.0 | 1.0 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.7 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.1 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.0 | 3.8 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 1.4 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 41.1 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 5.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.3 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.3 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 2.5 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 3.7 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 2.7 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.4 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.4 | GO:0061641 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0070327 | aromatic amino acid transport(GO:0015801) thyroid hormone transport(GO:0070327) |
| 0.0 | 0.0 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.0 | 0.2 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 11.0 | GO:0032584 | growth cone membrane(GO:0032584) |
| 1.4 | 9.5 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 1.2 | 4.6 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 1.1 | 6.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.7 | 2.0 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.6 | 3.7 | GO:0033063 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.6 | 12.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.5 | 1.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.5 | 26.6 | GO:0031430 | M band(GO:0031430) |
| 0.4 | 1.6 | GO:0071546 | pi-body(GO:0071546) |
| 0.3 | 2.7 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.3 | 3.7 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.2 | 13.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 1.0 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.2 | 0.9 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.2 | 6.0 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.2 | 45.1 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.2 | 11.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 2.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 1.9 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 6.4 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 1.8 | GO:0097431 | pericentriolar material(GO:0000242) mitotic spindle pole(GO:0097431) |
| 0.1 | 7.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 8.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 3.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 0.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.9 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 1.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.0 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.9 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 2.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 7.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 3.0 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 1.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 3.6 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 2.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 7.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 5.4 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 14.9 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 13.6 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 1.9 | 9.5 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 1.8 | 7.4 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 1.2 | 11.0 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.5 | 3.7 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.4 | 26.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.4 | 2.0 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.3 | 3.8 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.3 | 5.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.3 | 1.3 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.2 | 15.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 7.1 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.2 | 4.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.2 | 1.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.2 | 3.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 42.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.2 | 2.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.6 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 2.9 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 6.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 4.7 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.6 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.1 | 13.1 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 5.2 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 1.0 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 2.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 6.2 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 1.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 5.9 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 2.0 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.0 | 1.0 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 2.1 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 1.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 1.4 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.0 | 0.8 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 7.7 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 1.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.8 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 1.9 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 1.3 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 2.5 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 3.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 11.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 3.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 5.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 2.0 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.0 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 4.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 8.7 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 2.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 2.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.0 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 2.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.9 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.8 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 1.1 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 1.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |