Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for EOMES
Z-value: 6.80
Transcription factors associated with EOMES
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EOMES
|
ENSG00000163508.13 | EOMES |
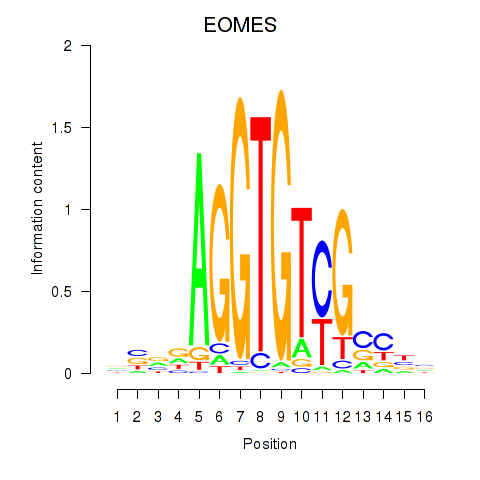
Activity profile of EOMES motif
Sorted Z-values of EOMES motif
Network of associatons between targets according to the STRING database.
First level regulatory network of EOMES
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.0 | 35.2 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 6.0 | 23.9 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 4.9 | 14.6 | GO:0014810 | positive regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014810) |
| 4.5 | 27.0 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 4.4 | 13.1 | GO:0046080 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 4.1 | 24.8 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 3.5 | 10.4 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) mitochondrial membrane fission(GO:0090149) |
| 3.2 | 9.7 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 3.2 | 9.5 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 2.8 | 14.2 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 2.7 | 15.9 | GO:0015853 | adenine transport(GO:0015853) |
| 2.5 | 7.6 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 2.5 | 34.9 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 2.4 | 12.0 | GO:0035549 | positive regulation of interferon-beta secretion(GO:0035549) |
| 2.4 | 11.8 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 2.3 | 7.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 2.2 | 10.8 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 2.1 | 27.6 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 2.1 | 8.4 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 2.0 | 18.3 | GO:0006167 | AMP biosynthetic process(GO:0006167) 'de novo' IMP biosynthetic process(GO:0006189) |
| 2.0 | 30.3 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 2.0 | 9.8 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 1.8 | 10.9 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 1.6 | 6.5 | GO:1903984 | rhythmic synaptic transmission(GO:0060024) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 1.6 | 4.8 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 1.6 | 6.3 | GO:1903755 | regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 1.5 | 6.2 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 1.5 | 7.5 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 1.5 | 29.1 | GO:0000022 | mitotic spindle elongation(GO:0000022) |
| 1.5 | 5.8 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 1.4 | 9.8 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 1.4 | 6.8 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 1.3 | 22.6 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 1.3 | 4.0 | GO:0001188 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 1.3 | 6.4 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 1.2 | 16.2 | GO:0006983 | ER overload response(GO:0006983) |
| 1.2 | 4.8 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 1.2 | 3.6 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 1.2 | 3.5 | GO:2000832 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 1.1 | 5.6 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 1.1 | 4.3 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 1.1 | 6.4 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 1.1 | 4.2 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 1.0 | 9.4 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 1.0 | 3.1 | GO:0060739 | mesenchymal-epithelial cell signaling involved in prostate gland development(GO:0060739) |
| 1.0 | 3.0 | GO:0051231 | spindle elongation(GO:0051231) |
| 1.0 | 13.3 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.9 | 2.8 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 0.9 | 14.1 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.9 | 5.3 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.9 | 5.3 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.9 | 4.3 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.8 | 8.4 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.8 | 4.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.8 | 9.1 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.8 | 8.2 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.8 | 6.6 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.8 | 1.6 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.8 | 5.7 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.8 | 4.8 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.7 | 7.3 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.7 | 6.4 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.7 | 25.0 | GO:0034080 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.7 | 3.5 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.7 | 6.9 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.7 | 8.6 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.7 | 4.6 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.7 | 3.3 | GO:0012502 | induction of programmed cell death(GO:0012502) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) |
| 0.6 | 3.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.6 | 1.9 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.6 | 9.1 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.6 | 2.4 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.6 | 1.1 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.5 | 1.6 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.5 | 10.8 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.5 | 2.0 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.5 | 4.0 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.5 | 7.8 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.5 | 1.4 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.4 | 2.6 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.4 | 2.2 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.4 | 3.6 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.4 | 12.9 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.4 | 2.7 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.4 | 10.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.4 | 3.8 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.4 | 1.1 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 0.4 | 16.2 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.4 | 9.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.4 | 5.1 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.4 | 7.6 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.4 | 1.8 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.4 | 23.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.4 | 2.8 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.4 | 4.9 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.3 | 7.0 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.3 | 1.0 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.3 | 1.0 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.3 | 8.7 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.3 | 1.0 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.3 | 3.9 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.3 | 1.3 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.3 | 5.4 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.3 | 1.6 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.3 | 0.9 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.3 | 1.2 | GO:0072106 | regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) |
| 0.3 | 3.6 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.3 | 13.2 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.3 | 5.2 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.3 | 2.6 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.3 | 3.7 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.3 | 2.0 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.3 | 0.8 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.3 | 1.4 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.3 | 3.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.3 | 13.4 | GO:0006294 | nucleotide-excision repair, preincision complex assembly(GO:0006294) |
| 0.3 | 18.2 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.3 | 2.9 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.3 | 4.3 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.3 | 1.3 | GO:0050915 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) sensory perception of sour taste(GO:0050915) |
| 0.3 | 1.6 | GO:0061743 | motor learning(GO:0061743) |
| 0.3 | 9.3 | GO:2000816 | negative regulation of mitotic sister chromatid separation(GO:2000816) |
| 0.3 | 3.4 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.3 | 3.4 | GO:0046040 | IMP metabolic process(GO:0046040) |
| 0.3 | 1.8 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.3 | 3.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.3 | 9.6 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.3 | 6.8 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.2 | 3.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.2 | 9.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.2 | 5.9 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.2 | 6.5 | GO:0001825 | blastocyst formation(GO:0001825) |
| 0.2 | 3.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 0.7 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.2 | 0.9 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.2 | 6.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.2 | 0.7 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.2 | 3.0 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.2 | 4.8 | GO:0051639 | actin filament network formation(GO:0051639) actin crosslink formation(GO:0051764) |
| 0.2 | 2.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 7.1 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.2 | 7.9 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.2 | 1.4 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.2 | 1.2 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.2 | 2.7 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.2 | 3.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.2 | 2.0 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.2 | 0.9 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.2 | 2.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 1.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 1.9 | GO:0055026 | negative regulation of cardiac muscle tissue development(GO:0055026) |
| 0.1 | 14.8 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.1 | 2.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 13.8 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 2.8 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.1 | 7.2 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 3.8 | GO:0036260 | 7-methylguanosine RNA capping(GO:0009452) RNA capping(GO:0036260) |
| 0.1 | 1.2 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.1 | 8.0 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 | 10.4 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.1 | 2.7 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 14.7 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 2.7 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.1 | 1.0 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 3.7 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.1 | 6.1 | GO:0032392 | DNA geometric change(GO:0032392) |
| 0.1 | 2.1 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 6.7 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 0.8 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 8.1 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.1 | 8.9 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.1 | 2.1 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.1 | 1.1 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.1 | 1.8 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.1 | 6.5 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 2.6 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.1 | 2.4 | GO:0043928 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 8.5 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.3 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 1.5 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 4.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 4.5 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.1 | 5.5 | GO:0061053 | somite development(GO:0061053) |
| 0.1 | 0.2 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.1 | 0.5 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 | 0.2 | GO:1903248 | negative regulation of cardiac muscle adaptation(GO:0010616) regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) regulation of cellular amino acid biosynthetic process(GO:2000282) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.1 | 5.0 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 3.1 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.1 | 0.4 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 2.4 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.1 | 2.8 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.5 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.9 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 | 5.5 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 2.7 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 1.2 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.0 | 1.3 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.1 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.0 | 1.0 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.1 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.0 | 2.9 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.3 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 1.0 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 4.5 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.9 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.4 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 2.0 | GO:1903169 | regulation of calcium ion transmembrane transport(GO:1903169) |
| 0.0 | 0.6 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 0.8 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.0 | 0.4 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.2 | GO:0048240 | sperm capacitation(GO:0048240) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 30.3 | GO:0000796 | condensin complex(GO:0000796) |
| 3.5 | 31.6 | GO:0097452 | GAIT complex(GO:0097452) |
| 1.8 | 24.0 | GO:0042555 | MCM complex(GO:0042555) |
| 1.8 | 9.1 | GO:0089701 | U2AF(GO:0089701) |
| 1.8 | 7.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 1.8 | 7.1 | GO:0071942 | XPC complex(GO:0071942) |
| 1.7 | 7.0 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 1.7 | 6.8 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 1.6 | 23.0 | GO:0070938 | contractile ring(GO:0070938) |
| 1.6 | 6.4 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 1.6 | 25.6 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 1.6 | 6.3 | GO:1990356 | sumoylated E2 ligase complex(GO:1990356) |
| 1.5 | 14.5 | GO:0097255 | R2TP complex(GO:0097255) |
| 1.3 | 27.0 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 1.3 | 13.0 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 1.3 | 14.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 1.2 | 4.8 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 1.2 | 7.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 1.1 | 1.1 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 1.1 | 10.9 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 1.0 | 23.1 | GO:0046930 | pore complex(GO:0046930) |
| 0.9 | 7.7 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.8 | 5.6 | GO:0031415 | NatA complex(GO:0031415) |
| 0.8 | 12.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.8 | 8.6 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.7 | 3.6 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.7 | 6.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.6 | 9.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.6 | 7.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.6 | 3.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.5 | 3.3 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.5 | 8.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.5 | 4.8 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.5 | 6.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.5 | 3.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.5 | 9.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.5 | 14.2 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.5 | 6.5 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.5 | 2.0 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.5 | 10.8 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.5 | 9.1 | GO:0036038 | MKS complex(GO:0036038) |
| 0.5 | 8.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.5 | 11.8 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.4 | 2.2 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.4 | 10.5 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.4 | 37.0 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.4 | 3.7 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.4 | 23.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.4 | 2.8 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.3 | 6.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.3 | 9.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.3 | 4.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.3 | 5.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.3 | 2.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.3 | 1.4 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.3 | 1.6 | GO:0070187 | telosome(GO:0070187) |
| 0.3 | 24.1 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.3 | 5.6 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.3 | 6.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 1.0 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 16.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 7.1 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.2 | 6.6 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.2 | 6.8 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.2 | 16.5 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.2 | 5.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 2.0 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 3.9 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.2 | 4.3 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.2 | 10.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 2.0 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.2 | 3.2 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.2 | 3.9 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 14.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 3.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 6.9 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 3.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 5.0 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.1 | 4.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 2.2 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 9.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 3.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 4.8 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 1.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 14.8 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 3.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.4 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 10.7 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 1.8 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 2.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 1.2 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 6.2 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 0.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 7.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 2.4 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 6.3 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 5.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 3.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.1 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 2.6 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 4.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.1 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 7.3 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.2 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 1.0 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 2.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.1 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 4.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 15.7 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.4 | GO:0031941 | filamentous actin(GO:0031941) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 13.1 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 3.7 | 11.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 3.2 | 9.7 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 2.9 | 14.6 | GO:0050610 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 2.9 | 23.1 | GO:0015288 | porin activity(GO:0015288) |
| 2.7 | 10.9 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 2.7 | 13.5 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 2.7 | 24.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 2.6 | 7.7 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 2.6 | 25.6 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 2.4 | 9.5 | GO:0004146 | dihydrofolate reductase activity(GO:0004146) |
| 2.2 | 6.5 | GO:0051717 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) |
| 2.1 | 6.2 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 1.9 | 13.0 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 1.8 | 7.1 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 1.8 | 5.3 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 1.6 | 6.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 1.5 | 11.8 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 1.4 | 4.3 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 1.3 | 6.6 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 1.3 | 3.8 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 1.2 | 16.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 1.2 | 10.9 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 1.2 | 7.0 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 1.2 | 4.6 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 1.1 | 3.4 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 1.1 | 23.9 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 1.0 | 4.0 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 1.0 | 7.7 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.9 | 2.8 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.9 | 31.6 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.9 | 3.6 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.9 | 6.8 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.8 | 6.8 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.8 | 38.2 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.8 | 12.7 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.8 | 2.3 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.7 | 3.7 | GO:0030620 | U1 snRNA binding(GO:0030619) U2 snRNA binding(GO:0030620) |
| 0.7 | 1.3 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.6 | 3.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.6 | 6.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.6 | 3.9 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.6 | 4.5 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.5 | 10.4 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.5 | 23.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.5 | 5.4 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.5 | 4.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.5 | 32.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.5 | 4.8 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.5 | 9.1 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.5 | 9.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.5 | 10.8 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.5 | 1.8 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.5 | 22.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.4 | 2.6 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.4 | 5.3 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.4 | 8.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.4 | 82.7 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.4 | 1.2 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.4 | 13.0 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.4 | 1.2 | GO:0009032 | thymidine phosphorylase activity(GO:0009032) pyrimidine-nucleoside phosphorylase activity(GO:0016154) |
| 0.4 | 1.2 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.4 | 3.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.4 | 3.8 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.4 | 2.2 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.4 | 3.7 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.3 | 3.5 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.3 | 2.7 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.3 | 1.3 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.3 | 1.3 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.3 | 10.7 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.3 | 12.8 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.3 | 17.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.3 | 11.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.3 | 8.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.3 | 7.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.3 | 1.2 | GO:0019976 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.3 | 2.9 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.3 | 0.8 | GO:0097258 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.3 | 3.8 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.3 | 7.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.3 | 6.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.3 | 3.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.3 | 5.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 2.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 6.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 28.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.2 | 3.5 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.2 | 5.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 7.6 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.2 | 2.4 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.2 | 3.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 3.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 5.0 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 2.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.2 | 13.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 1.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 1.2 | GO:0034617 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) tetrahydrobiopterin binding(GO:0034617) |
| 0.1 | 1.0 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.1 | 7.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 2.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 4.3 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 1.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 1.0 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.4 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 4.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 11.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 3.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 2.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 1.5 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 3.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 10.1 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.1 | 18.4 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.1 | 7.7 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 3.0 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 1.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 2.1 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.1 | 0.2 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 2.0 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 6.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 0.9 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 10.5 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 2.8 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 7.5 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 1.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 11.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 1.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 1.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 1.1 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 1.1 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.9 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.9 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 4.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 1.6 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.5 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 3.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 6.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 4.1 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 1.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 4.0 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 2.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.8 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 1.0 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 1.9 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 0.5 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.4 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 1.6 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 3.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 2.6 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 30.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.7 | 45.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.6 | 38.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.6 | 27.0 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.5 | 16.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.4 | 54.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.3 | 12.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.3 | 6.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.3 | 25.1 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.2 | 8.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 3.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 3.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.2 | 10.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.2 | 10.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.2 | 12.5 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.2 | 4.8 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.2 | 4.9 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.2 | 13.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 5.6 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.2 | 2.7 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.2 | 2.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 4.6 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 11.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 0.8 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 1.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 3.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 3.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 1.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 1.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 3.3 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 3.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 2.5 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 1.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.1 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.3 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.7 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.2 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.1 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 24.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 1.2 | 18.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 1.1 | 25.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 1.0 | 25.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.7 | 14.7 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.6 | 12.0 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.6 | 12.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.5 | 13.9 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.5 | 10.8 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.4 | 4.9 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.4 | 14.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.4 | 10.8 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.4 | 8.7 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.4 | 9.3 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.4 | 17.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.4 | 14.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.4 | 8.4 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.4 | 10.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.4 | 8.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 6.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.3 | 3.0 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.3 | 9.3 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.3 | 8.9 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.3 | 8.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.3 | 23.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.3 | 6.6 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.3 | 5.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 7.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.3 | 7.1 | REACTOME GLOBAL GENOMIC NER GG NER | Genes involved in Global Genomic NER (GG-NER) |
| 0.3 | 3.0 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.2 | 23.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 6.7 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.2 | 5.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.2 | 5.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 1.6 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.2 | 3.6 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.2 | 5.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 14.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.2 | 3.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 1.9 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.2 | 6.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 18.0 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.2 | 2.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 9.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 4.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 14.5 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.1 | 2.9 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 6.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 3.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 1.4 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.1 | 7.0 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.1 | 5.7 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 4.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 7.1 | REACTOME ORC1 REMOVAL FROM CHROMATIN | Genes involved in Orc1 removal from chromatin |
| 0.1 | 10.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.0 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.9 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 2.9 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 3.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 2.0 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 3.4 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 3.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 2.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 5.5 | REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | Genes involved in Processing of Capped Intron-Containing Pre-mRNA |
| 0.1 | 3.9 | REACTOME PI METABOLISM | Genes involved in PI Metabolism |
| 0.1 | 1.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 2.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 2.7 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 1.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 3.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.9 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 1.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 1.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 3.0 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 1.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 3.1 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.0 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |