Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
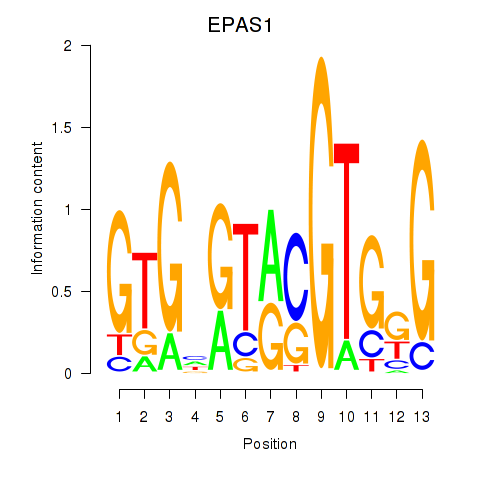
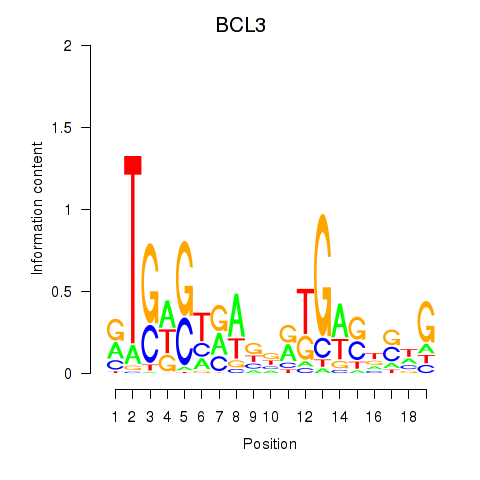
Results for EPAS1_BCL3
Z-value: 13.25
Transcription factors associated with EPAS1_BCL3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EPAS1
|
ENSG00000116016.14 | EPAS1 |
|
BCL3
|
ENSG00000069399.15 | BCL3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EPAS1 | hg38_v1_chr2_+_46297397_46297414 | 0.52 | 1.2e-16 | Click! |
| BCL3 | hg38_v1_chr19_+_44748673_44748749 | -0.20 | 2.8e-03 | Click! |
Activity profile of EPAS1_BCL3 motif
Sorted Z-values of EPAS1_BCL3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of EPAS1_BCL3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 33.8 | 202.5 | GO:0010734 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 17.6 | 282.2 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 15.6 | 109.4 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 15.5 | 77.5 | GO:0034443 | negative regulation of lipoprotein oxidation(GO:0034443) |
| 14.4 | 71.9 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 14.2 | 42.7 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 11.1 | 77.7 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 11.0 | 33.0 | GO:0071206 | establishment of protein localization to juxtaparanode region of axon(GO:0071206) |
| 10.0 | 40.1 | GO:0042488 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 9.4 | 66.1 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 9.1 | 45.6 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 9.1 | 81.5 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 9.0 | 45.2 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 9.0 | 45.1 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 8.6 | 25.7 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 8.3 | 58.4 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 8.3 | 66.5 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 8.1 | 24.3 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 8.0 | 23.9 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 7.9 | 31.6 | GO:0072302 | posterior mesonephric tubule development(GO:0072166) negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 7.3 | 44.0 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 7.3 | 50.9 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 7.0 | 21.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 7.0 | 21.1 | GO:0018874 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 6.9 | 20.7 | GO:0048687 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 6.3 | 18.9 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 6.2 | 18.7 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 6.2 | 18.6 | GO:0009183 | ADP biosynthetic process(GO:0006172) purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 6.1 | 24.6 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 6.0 | 65.5 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 5.8 | 17.3 | GO:0071464 | cellular response to hydrostatic pressure(GO:0071464) |
| 5.6 | 16.7 | GO:1903977 | positive regulation of glial cell migration(GO:1903977) |
| 5.4 | 16.3 | GO:0051939 | gamma-aminobutyric acid import(GO:0051939) |
| 5.3 | 52.9 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 5.2 | 15.5 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 5.0 | 35.3 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 5.0 | 20.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 4.7 | 14.1 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 4.7 | 14.0 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 4.6 | 9.2 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 4.5 | 18.0 | GO:0021759 | globus pallidus development(GO:0021759) |
| 4.3 | 34.4 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 4.3 | 12.9 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 4.3 | 21.3 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 4.1 | 12.4 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 4.1 | 81.8 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 4.0 | 12.0 | GO:1990764 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 3.9 | 11.8 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 3.9 | 15.5 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 3.9 | 7.7 | GO:0050955 | thermoception(GO:0050955) |
| 3.8 | 11.5 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 3.8 | 11.4 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 3.8 | 11.4 | GO:1905075 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 3.8 | 11.3 | GO:2000722 | umbilical cord morphogenesis(GO:0036304) umbilical cord development(GO:0061027) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) |
| 3.7 | 7.4 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 3.6 | 14.5 | GO:1902023 | L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 3.5 | 10.4 | GO:0042938 | dipeptide transport(GO:0042938) |
| 3.4 | 147.9 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 3.4 | 13.7 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 3.4 | 10.1 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 3.4 | 94.0 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 3.4 | 20.1 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 3.3 | 13.3 | GO:0030035 | microspike assembly(GO:0030035) phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 3.2 | 28.6 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 3.1 | 9.4 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 3.1 | 9.4 | GO:0061056 | sclerotome development(GO:0061056) |
| 3.1 | 18.7 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 3.0 | 9.1 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 3.0 | 50.4 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 3.0 | 65.0 | GO:0002021 | response to dietary excess(GO:0002021) |
| 2.9 | 49.5 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 2.9 | 14.3 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 2.8 | 11.3 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 2.8 | 8.4 | GO:0019836 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 2.8 | 14.0 | GO:1902612 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 2.8 | 8.4 | GO:2001302 | cellular response to interleukin-13(GO:0035963) regulation of engulfment of apoptotic cell(GO:1901074) lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 2.8 | 22.2 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 2.7 | 38.5 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 2.7 | 43.5 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 2.7 | 13.6 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 2.7 | 8.0 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 2.7 | 10.7 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 2.7 | 5.3 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 2.6 | 23.5 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 2.6 | 7.8 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 2.6 | 20.9 | GO:0043308 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) regulation of histamine secretion by mast cell(GO:1903593) |
| 2.6 | 12.8 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 2.5 | 72.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 2.5 | 5.0 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 2.5 | 22.2 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 2.5 | 4.9 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 2.4 | 7.3 | GO:0070845 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 2.4 | 38.6 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 2.4 | 16.8 | GO:0071279 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) cellular response to cobalt ion(GO:0071279) |
| 2.4 | 26.1 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 2.4 | 11.8 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 2.4 | 9.4 | GO:2000327 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 2.4 | 25.9 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 2.3 | 7.0 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 2.3 | 9.3 | GO:0033602 | gamma-aminobutyric acid biosynthetic process(GO:0009449) gamma-aminobutyric acid catabolic process(GO:0009450) negative regulation of gamma-aminobutyric acid secretion(GO:0014053) negative regulation of dopamine secretion(GO:0033602) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 2.3 | 46.2 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 2.3 | 25.2 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 2.2 | 9.0 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 2.2 | 15.5 | GO:0060424 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 2.2 | 6.6 | GO:0019878 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 2.2 | 17.7 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 2.2 | 11.0 | GO:1902943 | regulation of voltage-gated chloride channel activity(GO:1902941) positive regulation of voltage-gated chloride channel activity(GO:1902943) |
| 2.1 | 36.2 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 2.1 | 8.4 | GO:0010157 | response to chlorate(GO:0010157) |
| 2.1 | 4.2 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 2.1 | 10.4 | GO:0030070 | insulin processing(GO:0030070) |
| 2.1 | 31.3 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 2.1 | 6.2 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 2.1 | 6.2 | GO:0060994 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm development(GO:0048389) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) pattern specification involved in mesonephros development(GO:0061227) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in kidney development(GO:0072098) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 2.1 | 22.6 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 2.0 | 8.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 2.0 | 4.0 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 2.0 | 12.0 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 2.0 | 28.0 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 2.0 | 10.0 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 2.0 | 19.9 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 2.0 | 9.8 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 1.9 | 7.8 | GO:1904588 | cellular response to glycoprotein(GO:1904588) cellular response to thyrotropin-releasing hormone(GO:1905229) |
| 1.9 | 36.5 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 1.9 | 38.4 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 1.9 | 11.5 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 1.9 | 28.6 | GO:0035878 | nail development(GO:0035878) |
| 1.9 | 7.6 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 1.9 | 11.4 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 1.9 | 30.3 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 1.9 | 5.6 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 1.9 | 24.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 1.9 | 7.4 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 1.8 | 29.2 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 1.8 | 7.3 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 1.8 | 5.5 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 1.8 | 16.4 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 1.8 | 5.4 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 1.8 | 91.7 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 1.8 | 8.8 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 1.7 | 17.3 | GO:0006554 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 1.7 | 6.9 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) protein K29-linked ubiquitination(GO:0035519) |
| 1.7 | 8.5 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 1.7 | 6.8 | GO:1900145 | regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900175) |
| 1.7 | 6.8 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 1.6 | 27.8 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 1.6 | 81.5 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 1.6 | 3.2 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 1.6 | 9.5 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 1.6 | 6.3 | GO:0097283 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 1.6 | 4.7 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 1.5 | 12.3 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 1.5 | 6.1 | GO:0035938 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 1.5 | 18.4 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 1.5 | 6.1 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 1.5 | 4.6 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 1.5 | 10.7 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 1.5 | 4.6 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 1.5 | 9.0 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 1.5 | 10.4 | GO:0042428 | serotonin metabolic process(GO:0042428) |
| 1.5 | 5.9 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 1.5 | 36.8 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 1.5 | 29.3 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 1.4 | 10.1 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 1.4 | 12.9 | GO:0015886 | heme transport(GO:0015886) |
| 1.4 | 8.6 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 1.4 | 4.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 1.4 | 13.9 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 1.4 | 19.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 1.4 | 10.9 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 1.3 | 5.4 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) |
| 1.3 | 16.1 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 1.3 | 32.0 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 1.3 | 2.6 | GO:0099624 | atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 1.3 | 19.8 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 1.3 | 9.0 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 1.3 | 97.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 1.3 | 5.0 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 1.2 | 38.7 | GO:0048665 | neuron fate specification(GO:0048665) |
| 1.2 | 10.0 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 1.2 | 3.7 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 1.2 | 2.5 | GO:1902566 | regulation of eosinophil activation(GO:1902566) negative regulation of eosinophil activation(GO:1902567) |
| 1.2 | 29.6 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 1.2 | 6.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 1.2 | 85.2 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 1.2 | 13.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 1.2 | 3.6 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 1.2 | 1.2 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 1.2 | 21.2 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 1.2 | 17.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 1.2 | 22.2 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 1.2 | 4.6 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 1.2 | 1.2 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 1.1 | 3.4 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 1.1 | 17.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 1.1 | 14.6 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 1.1 | 5.6 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 1.1 | 7.7 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 1.1 | 12.9 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 1.1 | 9.6 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 1.1 | 5.4 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 1.1 | 8.5 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 1.1 | 19.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 1.1 | 61.5 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 1.1 | 12.6 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 1.1 | 53.6 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 1.0 | 7.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 1.0 | 14.5 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 1.0 | 2.1 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 1.0 | 14.1 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 1.0 | 4.0 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 1.0 | 13.9 | GO:0032196 | transposition(GO:0032196) |
| 1.0 | 6.9 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 1.0 | 3.9 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 1.0 | 2.9 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 1.0 | 4.8 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.9 | 7.5 | GO:0021681 | cerebellar granular layer development(GO:0021681) |
| 0.9 | 14.0 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.9 | 33.3 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.9 | 9.2 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.9 | 26.6 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.9 | 26.5 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.9 | 24.6 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.9 | 12.6 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.9 | 8.8 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.9 | 12.0 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.9 | 31.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.9 | 4.3 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.8 | 28.8 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.8 | 0.8 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.8 | 15.1 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.8 | 3.2 | GO:0001964 | startle response(GO:0001964) |
| 0.8 | 4.8 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.8 | 7.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.8 | 7.8 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.8 | 3.1 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.8 | 4.7 | GO:0032962 | regulation of inositol trisphosphate biosynthetic process(GO:0032960) positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.8 | 7.0 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.8 | 43.1 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.8 | 5.3 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.7 | 3.0 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.7 | 17.8 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.7 | 36.2 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.7 | 4.4 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.7 | 1.5 | GO:0001757 | somite specification(GO:0001757) |
| 0.7 | 13.2 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.7 | 2.9 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.7 | 15.4 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.7 | 40.3 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.7 | 5.8 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.7 | 2.2 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.7 | 9.3 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.7 | 18.6 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.7 | 2.8 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.7 | 31.0 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.7 | 19.0 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.7 | 51.3 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.7 | 11.1 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.7 | 10.3 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.7 | 6.2 | GO:0000050 | urea cycle(GO:0000050) |
| 0.7 | 1.3 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.7 | 2.0 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.7 | 15.1 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.6 | 14.3 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.6 | 18.2 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.6 | 17.4 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.6 | 4.4 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.6 | 8.8 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.6 | 7.5 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.6 | 28.1 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.6 | 2.4 | GO:0097491 | sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.6 | 17.9 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.6 | 16.6 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.6 | 14.0 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.6 | 6.3 | GO:0045986 | negative regulation of smooth muscle contraction(GO:0045986) |
| 0.6 | 4.0 | GO:0032026 | response to magnesium ion(GO:0032026) |
| 0.6 | 13.1 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.6 | 6.2 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.6 | 0.6 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.6 | 2.2 | GO:0035815 | positive regulation of renal sodium excretion(GO:0035815) |
| 0.6 | 6.6 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.5 | 3.3 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.5 | 4.4 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.5 | 5.5 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.5 | 10.2 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.5 | 10.2 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.5 | 6.4 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.5 | 29.6 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.5 | 43.8 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.5 | 5.7 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.5 | 1.6 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.5 | 6.7 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.5 | 6.6 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.5 | 9.7 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.5 | 4.1 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.5 | 5.0 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.5 | 18.0 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.5 | 3.9 | GO:1903859 | regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.5 | 4.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.5 | 13.7 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.5 | 3.3 | GO:1990822 | basic amino acid transmembrane transport(GO:1990822) |
| 0.5 | 22.2 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.5 | 24.9 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.5 | 13.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.5 | 1.8 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.5 | 11.8 | GO:0050779 | RNA destabilization(GO:0050779) |
| 0.5 | 6.8 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.4 | 11.7 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.4 | 4.9 | GO:0032094 | response to food(GO:0032094) |
| 0.4 | 11.1 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.4 | 1.8 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.4 | 2.2 | GO:0009624 | response to nematode(GO:0009624) |
| 0.4 | 12.7 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.4 | 6.9 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.4 | 10.0 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.4 | 6.0 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.4 | 35.0 | GO:0016079 | synaptic vesicle exocytosis(GO:0016079) |
| 0.4 | 8.4 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.4 | 5.0 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.4 | 20.1 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.4 | 2.5 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.4 | 16.7 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.4 | 4.7 | GO:0046500 | S-adenosylmethionine metabolic process(GO:0046500) |
| 0.4 | 1.9 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.4 | 4.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.4 | 5.8 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.4 | 41.0 | GO:0006024 | aminoglycan biosynthetic process(GO:0006023) glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.3 | 9.4 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.3 | 2.8 | GO:1902913 | positive regulation of melanocyte differentiation(GO:0045636) melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) positive regulation of neuroepithelial cell differentiation(GO:1902913) |
| 0.3 | 1.0 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.3 | 1.7 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.3 | 3.3 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.3 | 5.6 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.3 | 17.4 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.3 | 4.5 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.3 | 5.7 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.3 | 1.3 | GO:0019376 | galactolipid catabolic process(GO:0019376) |
| 0.3 | 1.6 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.3 | 20.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.3 | 0.9 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.3 | 3.9 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.3 | 6.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.3 | 1.7 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.3 | 2.5 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.3 | 7.8 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.3 | 1.9 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.3 | 0.8 | GO:0002818 | intracellular defense response(GO:0002818) |
| 0.3 | 6.0 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.3 | 1.6 | GO:0072498 | embryonic skeletal joint morphogenesis(GO:0060272) embryonic skeletal joint development(GO:0072498) |
| 0.3 | 0.5 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.3 | 5.0 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.3 | 5.9 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.3 | 7.4 | GO:0097502 | mannosylation(GO:0097502) |
| 0.3 | 9.5 | GO:0055001 | muscle cell development(GO:0055001) |
| 0.2 | 5.4 | GO:0043304 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.2 | 1.5 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.2 | 74.0 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.2 | 2.9 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.2 | 37.0 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.2 | 3.3 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.2 | 69.4 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.2 | 6.0 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.2 | 2.3 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.2 | 8.7 | GO:0006027 | aminoglycan catabolic process(GO:0006026) glycosaminoglycan catabolic process(GO:0006027) |
| 0.2 | 5.7 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.2 | 12.4 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.2 | 6.5 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.2 | 1.5 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.2 | 9.6 | GO:0006735 | NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.2 | 16.0 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.2 | 3.5 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.2 | 2.2 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.2 | 0.6 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.2 | 14.6 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.2 | 3.1 | GO:0007616 | long-term memory(GO:0007616) |
| 0.2 | 0.3 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.2 | 0.3 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.2 | 1.0 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.2 | 10.9 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.2 | 3.4 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.2 | 1.2 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.2 | 16.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.2 | 1.8 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 3.4 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 3.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 12.9 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 2.6 | GO:0048536 | spleen development(GO:0048536) |
| 0.1 | 2.9 | GO:0071420 | response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 0.1 | 1.9 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 3.6 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 8.3 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 11.7 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 10.6 | GO:0007626 | locomotory behavior(GO:0007626) |
| 0.1 | 5.5 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 0.2 | GO:0035948 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.1 | 1.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 6.9 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 1.1 | GO:0002544 | chronic inflammatory response(GO:0002544) |
| 0.1 | 0.6 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 0.6 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 1.7 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.1 | 12.5 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.1 | 2.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 0.4 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.1 | 7.5 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.1 | 0.8 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 2.2 | GO:0034383 | low-density lipoprotein particle clearance(GO:0034383) |
| 0.1 | 5.1 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.1 | 9.0 | GO:0031124 | mRNA 3'-end processing(GO:0031124) |
| 0.1 | 1.9 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 4.0 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 3.3 | GO:0051591 | response to cAMP(GO:0051591) |
| 0.1 | 4.0 | GO:0007259 | JAK-STAT cascade(GO:0007259) STAT cascade(GO:0097696) |
| 0.0 | 0.4 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 2.0 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.3 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 1.2 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) inositol lipid-mediated signaling(GO:0048017) |
| 0.0 | 0.6 | GO:0006582 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.0 | 1.0 | GO:0016049 | cell growth(GO:0016049) |
| 0.0 | 0.7 | GO:0031295 | T cell costimulation(GO:0031295) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 55.6 | 278.0 | GO:0072534 | perineuronal net(GO:0072534) |
| 25.0 | 75.0 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 23.2 | 92.8 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 11.5 | 206.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 11.4 | 45.8 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 9.0 | 45.2 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 8.1 | 24.3 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 7.5 | 45.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 6.7 | 40.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 6.4 | 25.7 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 6.1 | 67.1 | GO:0043203 | axon hillock(GO:0043203) |
| 5.1 | 46.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 5.1 | 40.8 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 4.2 | 21.1 | GO:0001652 | granular component(GO:0001652) |
| 4.2 | 16.8 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 4.2 | 20.8 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 4.0 | 64.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 3.4 | 85.7 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 3.4 | 10.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 3.3 | 26.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 3.0 | 11.9 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 2.8 | 11.1 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 2.6 | 7.7 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 2.4 | 7.1 | GO:0098536 | deuterosome(GO:0098536) |
| 2.3 | 9.3 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 2.2 | 17.4 | GO:0034464 | BBSome(GO:0034464) |
| 2.2 | 10.8 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 2.1 | 14.6 | GO:0060091 | kinocilium(GO:0060091) |
| 2.0 | 63.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 1.9 | 34.4 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 1.9 | 7.6 | GO:0070876 | SOSS complex(GO:0070876) |
| 1.9 | 26.3 | GO:0097433 | dense body(GO:0097433) |
| 1.8 | 9.2 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 1.8 | 20.1 | GO:0061200 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 1.8 | 7.3 | GO:0071546 | pi-body(GO:0071546) |
| 1.7 | 24.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 1.7 | 61.0 | GO:0071565 | nBAF complex(GO:0071565) |
| 1.7 | 15.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 1.7 | 89.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 1.6 | 34.6 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 1.6 | 47.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 1.6 | 16.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 1.6 | 7.8 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 1.5 | 16.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 1.5 | 14.9 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 1.5 | 36.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 1.4 | 11.6 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 1.4 | 23.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 1.4 | 8.5 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 1.4 | 4.2 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 1.4 | 12.4 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 1.4 | 52.2 | GO:0030673 | axolemma(GO:0030673) |
| 1.3 | 35.6 | GO:0033268 | node of Ranvier(GO:0033268) |
| 1.2 | 23.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 1.2 | 7.4 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 1.2 | 21.8 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 1.2 | 18.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 1.2 | 12.0 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 1.1 | 65.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 1.1 | 7.8 | GO:0043196 | varicosity(GO:0043196) |
| 1.1 | 28.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 1.0 | 10.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 1.0 | 16.7 | GO:0044304 | main axon(GO:0044304) |
| 1.0 | 10.6 | GO:0016342 | catenin complex(GO:0016342) |
| 1.0 | 312.5 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 1.0 | 2.9 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 1.0 | 14.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.9 | 8.4 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.9 | 119.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.9 | 72.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.9 | 86.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.9 | 15.5 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.9 | 9.9 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.9 | 20.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.8 | 10.9 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.8 | 22.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.8 | 11.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.8 | 36.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.8 | 47.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.8 | 180.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.8 | 41.5 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.7 | 30.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.7 | 7.4 | GO:0032009 | early phagosome(GO:0032009) |
| 0.7 | 41.0 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.7 | 10.0 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.7 | 55.3 | GO:0098793 | presynapse(GO:0098793) |
| 0.7 | 14.0 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.7 | 4.0 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.7 | 23.6 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.7 | 105.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.7 | 31.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.6 | 6.9 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.6 | 9.0 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.6 | 7.0 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.6 | 37.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.6 | 5.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.6 | 11.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.5 | 4.9 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.5 | 41.7 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.5 | 3.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.5 | 4.4 | GO:0032589 | neuron projection membrane(GO:0032589) |
| 0.5 | 1.9 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.5 | 50.9 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.5 | 5.6 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.5 | 146.1 | GO:0030424 | axon(GO:0030424) |
| 0.4 | 41.8 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.4 | 1.8 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.4 | 4.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.4 | 3.0 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.4 | 3.9 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.4 | 1.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.4 | 1.2 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.4 | 189.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.4 | 15.4 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.3 | 2.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.3 | 1.7 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.3 | 4.0 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.3 | 5.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.3 | 10.0 | GO:0032421 | stereocilium bundle(GO:0032421) |
| 0.3 | 25.6 | GO:0042641 | actomyosin(GO:0042641) |
| 0.3 | 15.2 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.3 | 25.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.3 | 3.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.3 | 3.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.3 | 5.3 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.2 | 20.9 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.2 | 5.9 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.2 | 5.0 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.2 | 1.0 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 6.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 17.2 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.2 | 10.3 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.2 | 9.0 | GO:0043235 | receptor complex(GO:0043235) |
| 0.2 | 9.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.2 | 6.7 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.2 | 25.6 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.2 | 2.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 3.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 5.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.2 | 6.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.2 | 14.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.8 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.1 | 3.0 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 5.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 2.8 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 17.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 19.5 | GO:0030425 | dendrite(GO:0030425) |
| 0.1 | 7.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 7.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 1.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 5.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 7.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 30.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 3.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 101.1 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 1.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 4.1 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.7 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 11.9 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 3.3 | GO:0005929 | cilium(GO:0005929) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.6 | 50.2 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 11.3 | 45.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 10.8 | 43.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 10.2 | 10.2 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 10.2 | 81.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 8.5 | 273.6 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 7.5 | 37.4 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 7.2 | 43.0 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 6.5 | 26.1 | GO:0005151 | interleukin-1, Type II receptor binding(GO:0005151) |
| 6.4 | 19.2 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 6.3 | 31.6 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 6.1 | 55.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 5.9 | 35.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 5.8 | 17.5 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 5.7 | 17.1 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 5.6 | 83.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 5.2 | 25.8 | GO:0005119 | smoothened binding(GO:0005119) |
| 4.9 | 44.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 4.8 | 14.5 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 4.8 | 24.0 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 4.7 | 162.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 4.3 | 17.4 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 4.2 | 16.8 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 4.1 | 20.7 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 4.0 | 23.9 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 3.9 | 11.8 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 3.9 | 46.3 | GO:0008430 | selenium binding(GO:0008430) |
| 3.9 | 15.4 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 3.8 | 15.2 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 3.8 | 67.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 3.7 | 29.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 3.7 | 25.7 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 3.6 | 236.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 3.5 | 17.7 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 3.5 | 14.0 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 3.5 | 17.5 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 3.4 | 17.2 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 3.4 | 16.8 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 3.3 | 123.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 3.3 | 13.2 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 3.2 | 16.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 3.2 | 19.3 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 3.2 | 12.9 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 3.1 | 31.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 3.1 | 104.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 3.1 | 42.8 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 3.0 | 9.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 3.0 | 33.5 | GO:0008061 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 3.0 | 36.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 3.0 | 47.8 | GO:0031005 | filamin binding(GO:0031005) |
| 3.0 | 183.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 3.0 | 11.8 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 3.0 | 11.8 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 2.9 | 14.7 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 2.9 | 26.3 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 2.9 | 8.8 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 2.9 | 8.7 | GO:0030305 | heparanase activity(GO:0030305) |
| 2.9 | 20.0 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 2.8 | 37.0 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 2.8 | 11.3 | GO:0035939 | microsatellite binding(GO:0035939) |
| 2.8 | 11.3 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 2.8 | 8.4 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 2.8 | 24.9 | GO:0033691 | sialic acid binding(GO:0033691) |
| 2.7 | 21.9 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 2.7 | 8.1 | GO:0016422 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 2.7 | 13.3 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 2.6 | 2.6 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 2.5 | 20.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 2.5 | 7.4 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 2.5 | 7.4 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 2.4 | 33.8 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 2.4 | 7.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 2.4 | 12.0 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 2.4 | 21.3 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 2.4 | 7.1 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 2.3 | 7.0 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 2.3 | 9.3 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 2.3 | 11.5 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 2.3 | 52.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 2.3 | 9.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 2.2 | 9.0 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 2.2 | 9.0 | GO:0035473 | lipase binding(GO:0035473) |
| 2.2 | 8.8 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) |
| 2.2 | 4.4 | GO:0042806 | fucose binding(GO:0042806) |
| 2.2 | 15.1 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 2.1 | 21.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 2.1 | 34.9 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 2.0 | 53.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 2.0 | 10.0 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 2.0 | 15.9 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 2.0 | 6.0 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 2.0 | 47.3 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 2.0 | 17.6 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 1.9 | 11.5 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 1.9 | 7.6 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 1.9 | 16.8 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 1.9 | 11.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 1.8 | 10.7 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 1.7 | 10.4 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 1.7 | 33.1 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 1.7 | 6.9 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 1.7 | 34.2 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 1.7 | 10.2 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 1.7 | 6.7 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 1.7 | 6.7 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 1.7 | 16.6 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 1.7 | 6.6 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 1.6 | 16.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 1.6 | 20.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 1.6 | 25.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 1.6 | 61.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 1.5 | 32.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 1.5 | 31.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 1.5 | 77.0 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 1.5 | 67.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 1.4 | 4.3 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 1.4 | 11.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 1.4 | 11.4 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 1.4 | 8.5 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 1.4 | 5.5 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 1.4 | 6.8 | GO:0004803 | transposase activity(GO:0004803) |
| 1.3 | 9.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 1.3 | 46.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 1.3 | 13.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 1.3 | 12.7 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 1.2 | 18.6 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 1.2 | 6.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 1.2 | 6.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 1.2 | 45.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 1.2 | 2.4 | GO:0019863 | IgE binding(GO:0019863) |
| 1.2 | 6.0 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 1.2 | 28.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 1.2 | 43.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 1.2 | 18.9 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 1.2 | 4.7 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 1.1 | 9.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 1.1 | 3.4 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 1.1 | 13.6 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 1.1 | 10.0 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 1.1 | 39.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 1.1 | 14.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 1.1 | 11.8 | GO:0036122 | BMP binding(GO:0036122) |
| 1.1 | 5.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 1.1 | 7.4 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 1.0 | 3.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 1.0 | 4.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 1.0 | 5.0 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 1.0 | 4.9 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 1.0 | 28.6 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 1.0 | 7.8 | GO:0038064 | collagen receptor activity(GO:0038064) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 1.0 | 23.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 1.0 | 11.8 | GO:0019864 | IgG binding(GO:0019864) |
| 1.0 | 19.5 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 1.0 | 7.8 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 1.0 | 18.4 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 1.0 | 11.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 1.0 | 2.9 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.9 | 14.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.9 | 3.7 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.9 | 16.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.9 | 3.6 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.9 | 17.9 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.9 | 4.4 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.9 | 4.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.9 | 41.0 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.9 | 10.4 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.9 | 32.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.8 | 16.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.8 | 4.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.8 | 10.0 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.8 | 9.7 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.8 | 12.0 | GO:0008517 | water transmembrane transporter activity(GO:0005372) folic acid transporter activity(GO:0008517) water channel activity(GO:0015250) |
| 0.8 | 4.0 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.8 | 7.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.8 | 30.1 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.8 | 92.7 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.8 | 6.9 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.8 | 7.6 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.7 | 18.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.7 | 104.6 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.7 | 8.0 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.7 | 86.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.7 | 7.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.7 | 15.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.7 | 8.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.7 | 23.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.7 | 10.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.7 | 31.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.7 | 14.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.7 | 3.9 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.6 | 33.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.6 | 1.9 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.6 | 7.6 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.6 | 1.9 | GO:0004339 | glucan 1,4-alpha-glucosidase activity(GO:0004339) |
| 0.6 | 9.9 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.6 | 3.0 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.6 | 3.6 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.6 | 27.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.6 | 10.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.6 | 21.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.6 | 8.7 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.6 | 8.1 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.6 | 2.8 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.6 | 16.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.5 | 6.0 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.5 | 4.7 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.5 | 11.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.5 | 5.9 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.5 | 4.7 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.5 | 9.0 | GO:0017136 | NAD-dependent histone deacetylase activity(GO:0017136) |
| 0.5 | 10.7 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.5 | 6.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.5 | 5.0 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.5 | 10.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.4 | 2.7 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.4 | 2.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.4 | 6.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.4 | 7.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.4 | 14.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.4 | 9.9 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.4 | 12.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.4 | 1.2 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.4 | 53.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.4 | 5.7 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.4 | 4.5 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.4 | 1.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.4 | 66.9 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.4 | 2.9 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.4 | 3.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.3 | 6.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.3 | 33.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.3 | 5.8 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.3 | 4.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.3 | 2.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.3 | 2.1 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.3 | 3.3 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.3 | 5.3 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.3 | 94.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.3 | 5.0 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.3 | 85.2 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.3 | 5.3 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.3 | 3.8 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.3 | 1.6 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.3 | 13.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 9.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 1.9 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.2 | 17.4 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.2 | 3.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.2 | 2.5 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 3.5 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.2 | 16.9 | GO:0005267 | potassium channel activity(GO:0005267) |
| 0.2 | 19.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 14.6 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.2 | 1.5 | GO:0043426 | MRF binding(GO:0043426) |
| 0.2 | 0.6 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.2 | 3.6 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.2 | 6.9 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.2 | 14.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.2 | 55.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.2 | 14.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.2 | 4.8 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.2 | 1.5 | GO:0015172 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.1 | 12.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 11.9 | GO:0005525 | GTP binding(GO:0005525) |
| 0.1 | 1.8 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.1 | 0.8 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 5.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 1.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 2.1 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 0.6 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 1.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 3.3 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 9.0 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 0.4 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 0.1 | 0.6 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 2.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 17.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 1.7 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 0.7 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 3.6 | GO:0005230 | extracellular ligand-gated ion channel activity(GO:0005230) |
| 0.1 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 1.0 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 1.9 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 4.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 2.9 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 1.0 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 1.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.4 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 45.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 1.8 | 70.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 1.5 | 28.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 1.4 | 93.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 1.3 | 142.3 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 1.3 | 12.6 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 1.0 | 39.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 1.0 | 49.7 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 1.0 | 3.0 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.9 | 120.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.9 | 32.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.9 | 20.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.8 | 50.6 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.8 | 21.4 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.8 | 50.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.8 | 17.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.8 | 45.1 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.8 | 4.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.8 | 18.6 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.8 | 17.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.7 | 23.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.7 | 7.8 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.7 | 5.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.7 | 7.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.7 | 21.3 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.7 | 41.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.7 | 3.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.6 | 20.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.6 | 5.0 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.6 | 22.0 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.5 | 13.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.5 | 20.5 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.5 | 8.8 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.5 | 8.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.5 | 13.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.5 | 29.6 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.4 | 7.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.4 | 17.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.4 | 4.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.4 | 24.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.4 | 9.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.4 | 14.0 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.4 | 78.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.4 | 18.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.4 | 7.7 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.4 | 14.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.4 | 22.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.4 | 9.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 9.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.3 | 13.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.3 | 16.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.3 | 20.1 | PID P73PATHWAY | p73 transcription factor network |
| 0.3 | 8.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.3 | 5.0 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.3 | 4.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.3 | 14.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 3.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 7.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 7.2 | ST ADRENERGIC | Adrenergic Pathway |
| 0.2 | 6.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.2 | 9.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.2 | 5.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.2 | 4.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 4.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 2.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 5.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 6.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 2.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 22.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.7 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 133.7 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 5.4 | 173.0 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 3.5 | 14.1 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 3.2 | 6.4 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 3.1 | 58.1 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 3.0 | 66.0 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 2.5 | 107.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 2.4 | 47.1 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 2.3 | 53.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 2.2 | 96.9 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 2.2 | 32.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 2.1 | 23.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 2.1 | 48.0 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 2.1 | 45.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 2.0 | 57.0 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 1.8 | 44.0 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 1.8 | 12.7 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 1.6 | 35.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 1.6 | 36.8 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 1.4 | 41.0 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 1.4 | 58.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 1.3 | 7.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 1.3 | 12.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 1.3 | 42.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 1.3 | 11.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 1.2 | 17.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 1.2 | 40.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 1.1 | 11.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 1.0 | 19.5 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 1.0 | 18.4 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 1.0 | 15.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 1.0 | 20.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 1.0 | 61.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 1.0 | 71.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 1.0 | 41.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 1.0 | 7.7 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 1.0 | 5.7 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.9 | 9.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.9 | 10.4 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.9 | 8.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.8 | 21.7 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.8 | 52.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.8 | 13.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.8 | 30.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.8 | 29.8 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.8 | 14.5 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.8 | 17.4 | REACTOME HEPARAN SULFATE HEPARIN HS GAG METABOLISM | Genes involved in Heparan sulfate/heparin (HS-GAG) metabolism |
| 0.8 | 75.3 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.8 | 37.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.7 | 7.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.7 | 12.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.7 | 12.0 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.7 | 72.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.7 | 17.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.7 | 20.0 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.7 | 10.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.7 | 4.6 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.7 | 7.8 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.6 | 7.8 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.6 | 14.3 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.6 | 17.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.6 | 48.9 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.6 | 2.3 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.6 | 4.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.6 | 41.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.6 | 14.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.6 | 22.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.6 | 17.0 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.6 | 3.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.5 | 14.6 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.5 | 8.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.5 | 3.4 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.5 | 17.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.4 | 13.9 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.4 | 9.0 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.4 | 84.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.4 | 4.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.4 | 6.1 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.4 | 14.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.4 | 7.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.4 | 12.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.4 | 6.0 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.4 | 4.5 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.4 | 10.7 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.4 | 13.1 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.4 | 4.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.3 | 2.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.3 | 6.6 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.3 | 7.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.3 | 7.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.3 | 6.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.3 | 10.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.3 | 13.2 | REACTOME SIGNALING BY FGFR MUTANTS | Genes involved in Signaling by FGFR mutants |
| 0.3 | 5.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.3 | 100.1 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.3 | 27.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 4.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 8.3 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.2 | 18.8 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 6.2 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.2 | 9.0 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.2 | 3.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 5.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.2 | 11.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 3.8 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.8 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.1 | 2.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 2.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.4 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 1.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 3.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 4.0 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.7 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.9 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.3 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.6 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 1.2 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |