Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
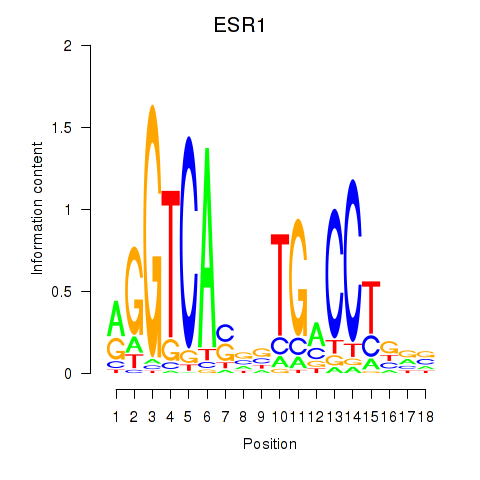
Results for ESR1
Z-value: 5.26
Transcription factors associated with ESR1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ESR1
|
ENSG00000091831.24 | ESR1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ESR1 | hg38_v1_chr6_+_151807674_151807692 | 0.26 | 9.7e-05 | Click! |
Activity profile of ESR1 motif
Sorted Z-values of ESR1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ESR1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_22758698 | 51.15 |
ENST00000390312.2
|
IGLV2-14
|
immunoglobulin lambda variable 2-14 |
| chr22_+_22906342 | 34.34 |
ENST00000390325.2
|
IGLC3
|
immunoglobulin lambda constant 3 (Kern-Oz+ marker) |
| chr22_+_22899481 | 32.92 |
ENST00000390322.2
|
IGLJ2
|
immunoglobulin lambda joining 2 |
| chr2_-_89100352 | 32.20 |
ENST00000479981.1
|
IGKV1-16
|
immunoglobulin kappa variable 1-16 |
| chr2_-_88992903 | 31.08 |
ENST00000495489.1
|
IGKV1-8
|
immunoglobulin kappa variable 1-8 |
| chr22_+_22904850 | 30.32 |
ENST00000390324.2
|
IGLJ3
|
immunoglobulin lambda joining 3 |
| chr22_+_22895368 | 29.80 |
ENST00000390321.2
|
IGLC1
|
immunoglobulin lambda constant 1 |
| chr22_+_22900976 | 28.90 |
ENST00000390323.2
|
IGLC2
|
immunoglobulin lambda constant 2 |
| chr2_-_89213917 | 28.51 |
ENST00000498435.1
|
IGKV1-27
|
immunoglobulin kappa variable 1-27 |
| chr14_-_105588322 | 23.05 |
ENST00000497872.4
ENST00000390539.2 |
IGHA2
|
immunoglobulin heavy constant alpha 2 (A2m marker) |
| chr22_+_22792485 | 22.73 |
ENST00000390314.2
|
IGLV2-11
|
immunoglobulin lambda variable 2-11 |
| chr22_+_22697789 | 22.61 |
ENST00000390306.2
|
IGLV2-23
|
immunoglobulin lambda variable 2-23 |
| chr22_+_22822658 | 20.96 |
ENST00000620395.2
|
IGLV2-8
|
immunoglobulin lambda variable 2-8 |
| chr14_-_105708627 | 20.11 |
ENST00000641837.1
ENST00000390547.3 |
IGHA1
|
immunoglobulin heavy constant alpha 1 |
| chr2_-_89010515 | 20.06 |
ENST00000493819.1
|
IGKV1-9
|
immunoglobulin kappa variable 1-9 |
| chr2_-_88979016 | 19.73 |
ENST00000390247.2
|
IGKV3-7
|
immunoglobulin kappa variable 3-7 (non-functional) |
| chr3_-_46464868 | 18.84 |
ENST00000417439.5
ENST00000231751.9 ENST00000431944.1 |
LTF
|
lactotransferrin |
| chr22_+_22704265 | 18.68 |
ENST00000390307.2
|
IGLV3-22
|
immunoglobulin lambda variable 3-22 |
| chr2_-_89117844 | 18.68 |
ENST00000490686.1
|
IGKV1-17
|
immunoglobulin kappa variable 1-17 |
| chr22_+_22357739 | 18.62 |
ENST00000390294.2
|
IGLV1-47
|
immunoglobulin lambda variable 1-47 |
| chr2_-_89297785 | 18.32 |
ENST00000465170.1
|
IGKV1-37
|
immunoglobulin kappa variable 1-37 (non-functional) |
| chr22_+_22594528 | 17.23 |
ENST00000390303.3
|
IGLV3-32
|
immunoglobulin lambda variable 3-32 (non-functional) |
| chr22_+_22686724 | 17.15 |
ENST00000390305.2
|
IGLV3-25
|
immunoglobulin lambda variable 3-25 |
| chr2_+_90038848 | 16.79 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20 |
| chr19_+_49335396 | 16.26 |
ENST00000598095.5
ENST00000426897.6 ENST00000323906.9 ENST00000535669.6 ENST00000597602.1 ENST00000595660.1 |
CD37
|
CD37 molecule |
| chr2_+_89862438 | 15.72 |
ENST00000448155.2
|
IGKV1D-39
|
immunoglobulin kappa variable 1D-39 |
| chr11_+_1870150 | 15.68 |
ENST00000429923.5
ENST00000418975.1 ENST00000406638.6 |
LSP1
|
lymphocyte specific protein 1 |
| chr2_-_88947820 | 15.38 |
ENST00000496168.1
|
IGKV1-5
|
immunoglobulin kappa variable 1-5 |
| chr11_+_1870252 | 14.92 |
ENST00000612798.4
|
LSP1
|
lymphocyte specific protein 1 |
| chr2_-_88966767 | 14.86 |
ENST00000464162.1
|
IGKV1-6
|
immunoglobulin kappa variable 1-6 |
| chr2_-_89268506 | 14.80 |
ENST00000473726.1
|
IGKV1-33
|
immunoglobulin kappa variable 1-33 |
| chr2_+_90154073 | 14.30 |
ENST00000611391.1
|
IGKV1D-13
|
immunoglobulin kappa variable 1D-13 |
| chr15_+_88639009 | 14.29 |
ENST00000306072.10
|
ISG20
|
interferon stimulated exonuclease gene 20 |
| chr22_+_22327298 | 13.81 |
ENST00000390291.2
|
IGLV1-50
|
immunoglobulin lambda variable 1-50 (non-functional) |
| chr15_+_88638947 | 13.75 |
ENST00000559876.2
|
ISG20
|
interferon stimulated exonuclease gene 20 |
| chr22_+_22588155 | 13.73 |
ENST00000390302.3
|
IGLV2-33
|
immunoglobulin lambda variable 2-33 (non-functional) |
| chr22_+_22380766 | 13.32 |
ENST00000390297.3
|
IGLV1-44
|
immunoglobulin lambda variable 1-44 |
| chr15_-_22160868 | 12.68 |
ENST00000604066.1
|
IGHV1OR15-1
|
immunoglobulin heavy variable 1/OR15-1 (non-functional) |
| chr19_-_45322867 | 12.16 |
ENST00000221476.4
|
CKM
|
creatine kinase, M-type |
| chr17_-_35880350 | 12.12 |
ENST00000605140.6
ENST00000651122.1 ENST00000603197.6 |
CCL5
|
C-C motif chemokine ligand 5 |
| chr12_+_111766887 | 12.04 |
ENST00000416293.7
ENST00000261733.7 |
ALDH2
|
aldehyde dehydrogenase 2 family member |
| chr22_+_22322452 | 11.71 |
ENST00000390290.3
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr14_-_94390650 | 11.67 |
ENST00000449399.7
ENST00000404814.8 |
SERPINA1
|
serpin family A member 1 |
| chr5_-_140633167 | 11.64 |
ENST00000302014.11
|
CD14
|
CD14 molecule |
| chr19_-_10334723 | 11.59 |
ENST00000592945.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr14_-_106269133 | 11.57 |
ENST00000390609.3
|
IGHV3-23
|
immunoglobulin heavy variable 3-23 |
| chrX_-_107775951 | 11.39 |
ENST00000315660.8
ENST00000372384.6 ENST00000502650.1 ENST00000506724.1 |
TSC22D3
|
TSC22 domain family member 3 |
| chr16_+_30183595 | 11.28 |
ENST00000219150.10
ENST00000570045.5 ENST00000565497.5 ENST00000570244.5 |
CORO1A
|
coronin 1A |
| chr14_-_106762576 | 11.26 |
ENST00000624687.1
|
IGHV1-69D
|
immunoglobulin heavy variable 1-69D |
| chr11_+_47248885 | 11.19 |
ENST00000395397.7
ENST00000405576.5 |
NR1H3
|
nuclear receptor subfamily 1 group H member 3 |
| chr11_+_47248924 | 10.72 |
ENST00000481889.6
ENST00000436778.5 ENST00000531660.5 ENST00000407404.5 |
NR1H3
|
nuclear receptor subfamily 1 group H member 3 |
| chr14_-_94390614 | 10.72 |
ENST00000553327.5
ENST00000556955.5 ENST00000557118.5 ENST00000440909.5 |
SERPINA1
|
serpin family A member 1 |
| chr14_-_94390667 | 10.62 |
ENST00000557492.5
ENST00000355814.8 ENST00000437397.5 ENST00000448921.5 ENST00000393088.8 |
SERPINA1
|
serpin family A member 1 |
| chr9_+_136980211 | 9.69 |
ENST00000444903.2
|
PTGDS
|
prostaglandin D2 synthase |
| chr14_-_106639589 | 9.41 |
ENST00000390630.3
|
IGHV4-61
|
immunoglobulin heavy variable 4-61 |
| chr19_+_18173804 | 9.41 |
ENST00000407280.4
|
IFI30
|
IFI30 lysosomal thiol reductase |
| chr4_+_674559 | 9.31 |
ENST00000511290.5
|
MYL5
|
myosin light chain 5 |
| chr19_-_6720641 | 9.07 |
ENST00000245907.11
|
C3
|
complement C3 |
| chr5_-_42825884 | 8.76 |
ENST00000506577.5
|
SELENOP
|
selenoprotein P |
| chr22_+_22409755 | 8.65 |
ENST00000390299.2
|
IGLV1-40
|
immunoglobulin lambda variable 1-40 |
| chr1_+_209675404 | 8.60 |
ENST00000367029.5
|
G0S2
|
G0/G1 switch 2 |
| chr12_-_14567714 | 8.59 |
ENST00000240617.10
|
PLBD1
|
phospholipase B domain containing 1 |
| chr13_-_46390042 | 8.42 |
ENST00000389908.7
|
RUBCNL
|
rubicon like autophagy enhancer |
| chr3_+_48446710 | 8.39 |
ENST00000635099.1
ENST00000346691.9 ENST00000320211.10 |
ATRIP
|
ATR interacting protein |
| chr19_-_13102848 | 7.92 |
ENST00000264824.5
|
LYL1
|
LYL1 basic helix-loop-helix family member |
| chr17_-_81911350 | 7.87 |
ENST00000570388.5
|
PCYT2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr3_+_186930759 | 7.83 |
ENST00000677292.1
ENST00000458216.5 |
ST6GAL1
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 1 |
| chr10_+_86968432 | 7.83 |
ENST00000416348.1
ENST00000372013.8 |
ADIRF
|
adipogenesis regulatory factor |
| chr3_+_46979659 | 7.75 |
ENST00000450053.8
|
NBEAL2
|
neurobeachin like 2 |
| chr10_+_112376193 | 7.51 |
ENST00000433418.6
ENST00000356116.6 ENST00000354273.5 |
ACSL5
|
acyl-CoA synthetase long chain family member 5 |
| chr11_-_47248835 | 7.49 |
ENST00000673604.1
ENST00000256997.9 |
ACP2
|
acid phosphatase 2, lysosomal |
| chr19_+_58278948 | 7.42 |
ENST00000621650.2
ENST00000591325.1 |
ZNF8
ZNF8-ERVK3-1
|
zinc finger protein 8 ZNF8-ERVK3-1 readthrough |
| chr11_-_615570 | 7.32 |
ENST00000649187.1
ENST00000647801.1 ENST00000397566.5 ENST00000397570.5 |
IRF7
|
interferon regulatory factor 7 |
| chr9_+_121207774 | 7.06 |
ENST00000373823.7
|
GSN
|
gelsolin |
| chr19_+_7903843 | 7.03 |
ENST00000397981.7
ENST00000397979.4 ENST00000397983.7 |
MAP2K7
|
mitogen-activated protein kinase kinase 7 |
| chr2_-_159904668 | 6.73 |
ENST00000504764.5
ENST00000505052.1 ENST00000263636.5 |
LY75-CD302
LY75
|
LY75-CD302 readthrough lymphocyte antigen 75 |
| chr2_-_219245389 | 6.71 |
ENST00000428427.1
ENST00000432839.1 ENST00000295759.12 ENST00000424620.1 |
GLB1L
|
galactosidase beta 1 like |
| chr1_-_111427731 | 6.70 |
ENST00000369732.4
|
OVGP1
|
oviductal glycoprotein 1 |
| chr11_-_47248789 | 6.65 |
ENST00000529444.7
ENST00000672787.1 ENST00000672073.1 ENST00000672636.2 ENST00000527256.7 |
ACP2
|
acid phosphatase 2, lysosomal |
| chr12_-_123972709 | 6.46 |
ENST00000545891.5
|
CCDC92
|
coiled-coil domain containing 92 |
| chr22_-_50578003 | 6.40 |
ENST00000405237.7
|
CPT1B
|
carnitine palmitoyltransferase 1B |
| chrX_-_107716401 | 6.39 |
ENST00000486554.1
ENST00000372390.8 |
TSC22D3
|
TSC22 domain family member 3 |
| chr1_+_27234612 | 6.38 |
ENST00000319394.8
ENST00000361771.7 |
WDTC1
|
WD and tetratricopeptide repeats 1 |
| chr16_+_1240698 | 6.31 |
ENST00000561736.2
ENST00000338844.8 ENST00000461509.6 |
TPSAB1
|
tryptase alpha/beta 1 |
| chr3_-_52454032 | 6.27 |
ENST00000232975.8
|
TNNC1
|
troponin C1, slow skeletal and cardiac type |
| chr3_+_186930518 | 6.25 |
ENST00000169298.8
ENST00000457772.6 ENST00000455441.5 ENST00000427315.5 |
ST6GAL1
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 1 |
| chr6_-_41735557 | 6.11 |
ENST00000373033.6
|
TFEB
|
transcription factor EB |
| chr16_-_28609992 | 6.09 |
ENST00000314752.11
|
SULT1A1
|
sulfotransferase family 1A member 1 |
| chr2_+_11539833 | 5.87 |
ENST00000263834.9
|
GREB1
|
growth regulating estrogen receptor binding 1 |
| chr6_+_31615215 | 5.84 |
ENST00000337917.11
ENST00000376059.8 |
AIF1
|
allograft inflammatory factor 1 |
| chr5_+_134905100 | 5.83 |
ENST00000512783.5
ENST00000254908.11 |
PCBD2
|
pterin-4 alpha-carbinolamine dehydratase 2 |
| chrY_-_1452882 | 5.77 |
ENST00000381317.9_PAR_Y
ENST00000381333.9_PAR_Y |
ASMTL
|
acetylserotonin O-methyltransferase like |
| chr1_-_23825402 | 5.75 |
ENST00000235958.4
ENST00000436439.6 ENST00000374490.8 |
HMGCL
|
3-hydroxy-3-methylglutaryl-CoA lyase |
| chr14_+_94110728 | 5.66 |
ENST00000616764.4
ENST00000618863.1 ENST00000611954.4 ENST00000618200.4 ENST00000621160.4 ENST00000555819.5 ENST00000620396.4 ENST00000612813.4 ENST00000620066.1 |
IFI27
|
interferon alpha inducible protein 27 |
| chr17_-_81911367 | 5.65 |
ENST00000538936.7
ENST00000538721.6 ENST00000573636.6 ENST00000571105.5 ENST00000576343.5 ENST00000572473.1 |
PCYT2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr6_-_41736239 | 5.64 |
ENST00000358871.6
|
TFEB
|
transcription factor EB |
| chr17_+_36064265 | 5.63 |
ENST00000616054.2
|
CCL18
|
C-C motif chemokine ligand 18 |
| chr22_-_50577915 | 5.62 |
ENST00000360719.6
ENST00000457250.5 |
CPT1B
|
carnitine palmitoyltransferase 1B |
| chr20_+_3820515 | 5.59 |
ENST00000246041.6
ENST00000379573.6 ENST00000379567.6 ENST00000455742.5 ENST00000615891.2 |
AP5S1
|
adaptor related protein complex 5 subunit sigma 1 |
| chr7_+_30028644 | 5.53 |
ENST00000440706.3
|
PLEKHA8
|
pleckstrin homology domain containing A8 |
| chr2_+_230225718 | 5.53 |
ENST00000420434.7
ENST00000392045.8 ENST00000417495.7 ENST00000343805.10 |
SP140
|
SP140 nuclear body protein |
| chr11_-_615921 | 5.50 |
ENST00000348655.11
ENST00000525445.6 ENST00000330243.9 |
IRF7
|
interferon regulatory factor 7 |
| chr11_-_60952559 | 5.43 |
ENST00000538739.2
|
SLC15A3
|
solute carrier family 15 member 3 |
| chr6_-_49744434 | 5.36 |
ENST00000433368.6
ENST00000354620.4 |
CRISP3
|
cysteine rich secretory protein 3 |
| chr17_-_81911200 | 5.32 |
ENST00000570391.5
|
PCYT2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr22_-_38755458 | 5.29 |
ENST00000405510.5
ENST00000433561.5 |
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr11_+_62419025 | 5.27 |
ENST00000278282.3
|
SCGB1A1
|
secretoglobin family 1A member 1 |
| chr17_-_41521719 | 5.10 |
ENST00000393976.6
|
KRT15
|
keratin 15 |
| chr20_+_49812697 | 5.05 |
ENST00000417961.5
|
SLC9A8
|
solute carrier family 9 member A8 |
| chr10_+_74826550 | 5.04 |
ENST00000649657.1
ENST00000372714.6 ENST00000649442.1 ENST00000648539.1 ENST00000647666.1 ENST00000648048.1 ENST00000287239.10 ENST00000649375.1 |
KAT6B
|
lysine acetyltransferase 6B |
| chr11_-_72752376 | 5.00 |
ENST00000393609.8
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr11_-_60952134 | 4.97 |
ENST00000679573.1
ENST00000681882.1 ENST00000681951.1 ENST00000227880.8 |
SLC15A3
|
solute carrier family 15 member 3 |
| chr15_+_28885955 | 4.95 |
ENST00000558402.5
ENST00000683413.1 ENST00000558330.5 |
APBA2
|
amyloid beta precursor protein binding family A member 2 |
| chr8_-_69833338 | 4.90 |
ENST00000524945.5
|
SLCO5A1
|
solute carrier organic anion transporter family member 5A1 |
| chr22_-_23754376 | 4.88 |
ENST00000398465.3
ENST00000248948.4 |
VPREB3
|
V-set pre-B cell surrogate light chain 3 |
| chr16_-_1230089 | 4.86 |
ENST00000612142.1
ENST00000606293.5 |
TPSB2
|
tryptase beta 2 |
| chr1_+_19664859 | 4.86 |
ENST00000289753.2
|
HTR6
|
5-hydroxytryptamine receptor 6 |
| chr1_+_19596960 | 4.85 |
ENST00000617872.4
ENST00000322753.7 ENST00000602662.1 |
MICOS10
NBL1
|
mitochondrial contact site and cristae organizing system subunit 10 NBL1, DAN family BMP antagonist |
| chrY_+_20575716 | 4.81 |
ENST00000361365.7
|
EIF1AY
|
eukaryotic translation initiation factor 1A Y-linked |
| chr11_+_43358908 | 4.77 |
ENST00000039989.9
ENST00000299240.10 |
TTC17
|
tetratricopeptide repeat domain 17 |
| chr20_+_49812818 | 4.75 |
ENST00000361573.3
|
SLC9A8
|
solute carrier family 9 member A8 |
| chr7_-_102543849 | 4.75 |
ENST00000644544.1
|
UPK3BL2
|
uroplakin 3B like 2 |
| chr11_-_60952067 | 4.69 |
ENST00000681275.1
|
SLC15A3
|
solute carrier family 15 member 3 |
| chr16_+_590200 | 4.68 |
ENST00000563109.1
|
RAB40C
|
RAB40C, member RAS oncogene family |
| chr22_-_23580223 | 4.68 |
ENST00000249053.3
ENST00000330377.3 ENST00000438703.1 |
IGLL1
|
immunoglobulin lambda like polypeptide 1 |
| chr3_-_15427497 | 4.62 |
ENST00000443029.5
ENST00000383789.9 ENST00000450816.6 ENST00000383790.8 |
METTL6
|
methyltransferase like 6 |
| chr6_-_2903300 | 4.60 |
ENST00000380698.5
|
SERPINB9
|
serpin family B member 9 |
| chr6_+_35297809 | 4.60 |
ENST00000316637.7
|
DEF6
|
DEF6 guanine nucleotide exchange factor |
| chr12_-_13103637 | 4.58 |
ENST00000545401.5
ENST00000432710.6 ENST00000351606.10 ENST00000651961.1 ENST00000542415.5 |
GSG1
|
germ cell associated 1 |
| chr6_-_90296824 | 4.56 |
ENST00000257749.9
|
BACH2
|
BTB domain and CNC homolog 2 |
| chr19_+_35268921 | 4.56 |
ENST00000222305.8
ENST00000343550.9 |
USF2
|
upstream transcription factor 2, c-fos interacting |
| chr15_+_81196871 | 4.55 |
ENST00000559383.5
ENST00000394660.6 ENST00000683961.1 |
IL16
|
interleukin 16 |
| chr1_+_9651723 | 4.55 |
ENST00000377346.9
ENST00000536656.5 ENST00000628140.2 |
PIK3CD
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta |
| chr1_+_220786853 | 4.54 |
ENST00000366910.10
|
MTARC1
|
mitochondrial amidoxime reducing component 1 |
| chr19_+_35533436 | 4.53 |
ENST00000222286.9
|
GAPDHS
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr19_+_35269065 | 4.50 |
ENST00000595068.5
ENST00000379134.7 ENST00000594064.5 ENST00000598058.1 |
USF2
|
upstream transcription factor 2, c-fos interacting |
| chr19_-_15331890 | 4.50 |
ENST00000594841.5
ENST00000601941.1 |
BRD4
|
bromodomain containing 4 |
| chr2_-_230225628 | 4.43 |
ENST00000540870.5
|
SP110
|
SP110 nuclear body protein |
| chr15_-_72783685 | 4.41 |
ENST00000456471.3
ENST00000311669.12 |
ADPGK
|
ADP dependent glucokinase |
| chr11_+_8682782 | 4.38 |
ENST00000531978.5
ENST00000524496.5 ENST00000532359.5 ENST00000314138.11 ENST00000530022.5 |
RPL27A
|
ribosomal protein L27a |
| chr5_-_154850570 | 4.37 |
ENST00000326080.10
ENST00000519501.5 ENST00000518651.5 ENST00000517938.5 ENST00000520461.1 |
FAXDC2
|
fatty acid hydroxylase domain containing 2 |
| chr6_+_29827817 | 4.36 |
ENST00000360323.11
ENST00000376818.7 ENST00000376815.3 |
HLA-G
|
major histocompatibility complex, class I, G |
| chr9_-_33264559 | 4.35 |
ENST00000473781.1
ENST00000379704.7 ENST00000488499.1 |
BAG1
|
BAG cochaperone 1 |
| chr19_+_2389761 | 4.30 |
ENST00000648592.1
|
TMPRSS9
|
transmembrane serine protease 9 |
| chr14_-_106360320 | 4.30 |
ENST00000390615.2
|
IGHV3-33
|
immunoglobulin heavy variable 3-33 |
| chr3_+_15427551 | 4.29 |
ENST00000396842.7
|
EAF1
|
ELL associated factor 1 |
| chr9_-_136205122 | 4.27 |
ENST00000371748.10
|
LHX3
|
LIM homeobox 3 |
| chr20_+_63981117 | 4.24 |
ENST00000266079.5
|
PRPF6
|
pre-mRNA processing factor 6 |
| chr16_+_85027735 | 4.21 |
ENST00000258180.7
ENST00000538274.5 |
KIAA0513
|
KIAA0513 |
| chr14_-_106005574 | 4.21 |
ENST00000390595.3
|
IGHV1-3
|
immunoglobulin heavy variable 1-3 |
| chr1_+_205227889 | 4.20 |
ENST00000358024.8
|
TMCC2
|
transmembrane and coiled-coil domain family 2 |
| chrY_+_20575792 | 4.20 |
ENST00000382772.3
|
EIF1AY
|
eukaryotic translation initiation factor 1A Y-linked |
| chr16_+_19718264 | 4.17 |
ENST00000564186.5
|
IQCK
|
IQ motif containing K |
| chr6_-_73452124 | 4.17 |
ENST00000680833.1
|
CGAS
|
cyclic GMP-AMP synthase |
| chr22_+_22195753 | 4.16 |
ENST00000390285.4
|
IGLV6-57
|
immunoglobulin lambda variable 6-57 |
| chr17_-_36388423 | 4.13 |
ENST00000610350.3
|
TBC1D3H
|
TBC1 domain family member 3H |
| chr22_-_36817001 | 4.13 |
ENST00000406910.6
ENST00000417718.7 |
PVALB
|
parvalbumin |
| chr8_+_11494367 | 4.12 |
ENST00000259089.9
ENST00000529894.1 |
BLK
|
BLK proto-oncogene, Src family tyrosine kinase |
| chr14_+_103529208 | 4.11 |
ENST00000299202.4
|
TRMT61A
|
tRNA methyltransferase 61A |
| chr16_+_10386049 | 4.11 |
ENST00000562527.5
ENST00000396559.5 ENST00000396560.6 ENST00000562102.5 ENST00000543967.5 ENST00000569939.5 ENST00000569900.5 |
ATF7IP2
|
activating transcription factor 7 interacting protein 2 |
| chr15_+_69452811 | 4.10 |
ENST00000357790.5
ENST00000260379.11 ENST00000560274.1 |
RPLP1
|
ribosomal protein lateral stalk subunit P1 |
| chr16_-_1826778 | 4.09 |
ENST00000569339.1
ENST00000397356.8 ENST00000455446.6 ENST00000397353.6 |
HAGH
|
hydroxyacylglutathione hydrolase |
| chr19_-_11529116 | 4.08 |
ENST00000592312.5
ENST00000590480.1 ENST00000585318.5 ENST00000270517.12 ENST00000252440.11 ENST00000417981.6 |
ECSIT
|
ECSIT signaling integrator |
| chr19_-_51009586 | 4.08 |
ENST00000594211.2
|
KLK9
|
kallikrein related peptidase 9 |
| chr22_-_42720813 | 4.06 |
ENST00000381278.4
|
A4GALT
|
alpha 1,4-galactosyltransferase (P blood group) |
| chr1_+_153776596 | 4.03 |
ENST00000458027.5
|
SLC27A3
|
solute carrier family 27 member 3 |
| chr19_-_4455292 | 4.03 |
ENST00000394765.7
ENST00000592515.1 |
UBXN6
|
UBX domain protein 6 |
| chr6_-_49744378 | 4.03 |
ENST00000371159.8
ENST00000263045.9 |
CRISP3
|
cysteine rich secretory protein 3 |
| chr12_-_123972824 | 4.02 |
ENST00000238156.8
ENST00000545037.1 |
CCDC92
|
coiled-coil domain containing 92 |
| chr11_-_35419542 | 4.00 |
ENST00000643305.1
ENST00000644351.1 ENST00000278379.9 ENST00000644779.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr21_+_39445824 | 3.94 |
ENST00000380637.7
ENST00000380634.5 ENST00000458295.5 ENST00000440288.6 ENST00000380631.5 |
SH3BGR
|
SH3 domain binding glutamate rich protein |
| chr1_+_12166978 | 3.93 |
ENST00000376259.7
ENST00000536782.2 |
TNFRSF1B
|
TNF receptor superfamily member 1B |
| chr12_+_113221429 | 3.93 |
ENST00000551096.5
ENST00000551099.5 ENST00000552897.5 ENST00000550785.5 ENST00000549279.1 ENST00000335509.11 |
TPCN1
|
two pore segment channel 1 |
| chr2_+_118942188 | 3.93 |
ENST00000327097.5
|
MARCO
|
macrophage receptor with collagenous structure |
| chr5_-_150086511 | 3.92 |
ENST00000675795.1
|
CSF1R
|
colony stimulating factor 1 receptor |
| chr3_-_184017863 | 3.91 |
ENST00000427120.6
ENST00000334444.11 ENST00000392579.6 ENST00000382494.6 ENST00000265586.10 ENST00000446941.2 |
ABCC5
|
ATP binding cassette subfamily C member 5 |
| chr11_+_8683201 | 3.89 |
ENST00000526562.5
ENST00000525981.1 |
RPL27A
|
ribosomal protein L27a |
| chr22_+_37639660 | 3.87 |
ENST00000649765.2
ENST00000451997.6 |
SH3BP1
ENSG00000285304.1
|
SH3 domain binding protein 1 novel protein |
| chr1_+_244969976 | 3.87 |
ENST00000366522.6
|
EFCAB2
|
EF-hand calcium binding domain 2 |
| chr1_+_54641806 | 3.86 |
ENST00000409996.5
|
MROH7
|
maestro heat like repeat family member 7 |
| chr16_+_85027761 | 3.79 |
ENST00000683363.1
|
KIAA0513
|
KIAA0513 |
| chr17_-_38192555 | 3.79 |
ENST00000620215.3
|
TBC1D3
|
TBC1 domain family member 3 |
| chr17_-_36439566 | 3.77 |
ENST00000620210.1
|
TBC1D3F
|
TBC1 domain family member 3F |
| chr17_+_38003976 | 3.76 |
ENST00000616101.4
|
TBC1D3D
|
TBC1 domain family member 3D |
| chr1_-_155188351 | 3.69 |
ENST00000462317.5
|
MUC1
|
mucin 1, cell surface associated |
| chr1_-_226737277 | 3.67 |
ENST00000272117.7
|
ITPKB
|
inositol-trisphosphate 3-kinase B |
| chr16_+_29459913 | 3.66 |
ENST00000360423.12
|
SULT1A4
|
sulfotransferase family 1A member 4 |
| chr8_+_102551583 | 3.63 |
ENST00000285402.4
|
ODF1
|
outer dense fiber of sperm tails 1 |
| chr17_+_37924415 | 3.62 |
ENST00000620247.1
|
TBC1D3K
|
TBC1 domain family member 3K |
| chr19_-_913182 | 3.62 |
ENST00000361574.10
ENST00000587975.2 |
R3HDM4
|
R3H domain containing 4 |
| chr3_-_14178569 | 3.61 |
ENST00000285021.12
|
XPC
|
XPC complex subunit, DNA damage recognition and repair factor |
| chr3_+_88059231 | 3.60 |
ENST00000636215.2
|
ZNF654
|
zinc finger protein 654 |
| chr17_-_37989048 | 3.59 |
ENST00000617678.2
ENST00000612727.5 |
TBC1D3L
|
TBC1 domain family member 3L |
| chr5_+_40679907 | 3.59 |
ENST00000302472.4
|
PTGER4
|
prostaglandin E receptor 4 |
| chr5_-_132011794 | 3.58 |
ENST00000650697.1
|
ACSL6
|
acyl-CoA synthetase long chain family member 6 |
| chr1_+_54641754 | 3.57 |
ENST00000339553.9
ENST00000421030.7 |
MROH7
|
maestro heat like repeat family member 7 |
| chr17_+_38127951 | 3.55 |
ENST00000621587.2
|
TBC1D3E
|
TBC1 domain family member 3E |
| chr10_-_98446878 | 3.54 |
ENST00000338546.9
ENST00000325103.10 ENST00000361490.9 |
HPS1
|
HPS1 biogenesis of lysosomal organelles complex 3 subunit 1 |
| chr17_+_36323884 | 3.54 |
ENST00000569055.2
|
TBC1D3G
|
TBC1 domain family member 3G |
| chr11_+_65639860 | 3.54 |
ENST00000527525.5
|
SIPA1
|
signal-induced proliferation-associated 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.3 | 28.0 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 7.3 | 21.9 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 4.8 | 43.3 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 4.7 | 18.8 | GO:1902732 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) positive regulation of chondrocyte proliferation(GO:1902732) |
| 3.7 | 22.4 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 3.0 | 9.1 | GO:0002445 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) |
| 2.7 | 393.4 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 2.3 | 11.6 | GO:0038123 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 2.3 | 9.1 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) late viral transcription(GO:0019086) |
| 2.2 | 6.7 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 2.2 | 17.8 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 2.1 | 6.3 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 1.9 | 11.3 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 1.7 | 6.8 | GO:1903627 | regulation of calcium-dependent ATPase activity(GO:1903610) negative regulation of calcium-dependent ATPase activity(GO:1903611) regulation of dUTP diphosphatase activity(GO:1903627) positive regulation of dUTP diphosphatase activity(GO:1903629) negative regulation of aminoacyl-tRNA ligase activity(GO:1903631) regulation of leucine-tRNA ligase activity(GO:1903633) negative regulation of leucine-tRNA ligase activity(GO:1903634) |
| 1.7 | 5.1 | GO:0070843 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 1.7 | 8.3 | GO:2000870 | oocyte growth(GO:0001555) regulation of progesterone secretion(GO:2000870) |
| 1.5 | 4.6 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 1.4 | 7.1 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 1.4 | 5.5 | GO:0035627 | ceramide transport(GO:0035627) |
| 1.3 | 5.0 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 1.2 | 4.8 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 1.2 | 6.0 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 1.2 | 3.6 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 1.2 | 5.9 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 1.2 | 8.2 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) |
| 1.2 | 5.8 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) |
| 1.1 | 2.3 | GO:0002461 | tolerance induction dependent upon immune response(GO:0002461) |
| 1.1 | 2.2 | GO:0002331 | pre-B cell differentiation(GO:0002329) pre-B cell allelic exclusion(GO:0002331) |
| 1.1 | 3.3 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) |
| 1.1 | 21.0 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 1.1 | 8.8 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 1.1 | 5.3 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 1.1 | 5.3 | GO:0010193 | response to ozone(GO:0010193) |
| 1.0 | 23.7 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 1.0 | 4.1 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 1.0 | 3.0 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 1.0 | 7.8 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 1.0 | 169.4 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.9 | 2.8 | GO:0032761 | negative regulation of T cell tolerance induction(GO:0002665) negative regulation of T cell anergy(GO:0002668) negative regulation of lymphocyte anergy(GO:0002912) regulation of lymphotoxin A production(GO:0032681) positive regulation of lymphotoxin A production(GO:0032761) regulation of lymphotoxin A biosynthetic process(GO:0043016) positive regulation of lymphotoxin A biosynthetic process(GO:0043017) |
| 0.9 | 7.5 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.9 | 5.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.9 | 3.5 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.9 | 15.9 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.9 | 4.4 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.9 | 3.5 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.9 | 2.6 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.9 | 2.6 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.8 | 3.4 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.8 | 3.4 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.8 | 12.2 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.8 | 7.0 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.8 | 10.8 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.8 | 3.8 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.7 | 3.0 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.7 | 5.8 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.7 | 2.2 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.7 | 4.3 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.7 | 2.8 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.7 | 2.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.7 | 2.7 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.7 | 2.0 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.7 | 7.9 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.6 | 7.1 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.6 | 3.2 | GO:1903238 | positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.6 | 2.4 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.6 | 1.8 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.6 | 3.5 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.6 | 3.5 | GO:0034127 | regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034127) |
| 0.6 | 6.4 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.6 | 1.7 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.5 | 1.6 | GO:0001757 | somite specification(GO:0001757) sclerotome development(GO:0061056) |
| 0.5 | 9.2 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.5 | 2.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.5 | 3.2 | GO:0075044 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.5 | 3.1 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.5 | 2.0 | GO:0009440 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.5 | 1.5 | GO:1903626 | positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.5 | 9.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.5 | 32.9 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.5 | 4.3 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.5 | 1.9 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.5 | 1.4 | GO:0071462 | cellular response to water stimulus(GO:0071462) cellular response to hydrostatic pressure(GO:0071464) |
| 0.5 | 1.4 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.4 | 2.7 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.4 | 2.2 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.4 | 3.9 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.4 | 15.6 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.4 | 1.7 | GO:0071110 | protein biotinylation(GO:0009305) histone biotinylation(GO:0071110) |
| 0.4 | 1.3 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.4 | 1.3 | GO:0009080 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.4 | 2.1 | GO:1901091 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.4 | 6.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.4 | 38.6 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.4 | 4.5 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.4 | 1.6 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.4 | 2.0 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.4 | 4.0 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.4 | 3.9 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.4 | 1.2 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.4 | 3.0 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.4 | 1.1 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.4 | 1.5 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.4 | 8.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.4 | 1.5 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) histone H3-K36 dimethylation(GO:0097676) |
| 0.4 | 8.0 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.4 | 5.8 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.4 | 2.5 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.4 | 1.8 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.4 | 15.5 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.4 | 30.2 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.3 | 3.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.3 | 1.4 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.3 | 3.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.3 | 3.1 | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) fat pad development(GO:0060613) positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) positive regulation of protein monoubiquitination(GO:1902527) |
| 0.3 | 1.7 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.3 | 2.2 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.3 | 3.1 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.3 | 2.2 | GO:0050666 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.3 | 3.4 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.3 | 3.0 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.3 | 1.2 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.3 | 2.4 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.3 | 0.9 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.3 | 0.9 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.3 | 3.2 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.3 | 92.1 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.3 | 2.8 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.3 | 2.2 | GO:0015886 | heme transport(GO:0015886) |
| 0.3 | 3.6 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.3 | 7.8 | GO:0030220 | platelet formation(GO:0030220) |
| 0.3 | 1.4 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.3 | 4.0 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.3 | 2.3 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.2 | 2.0 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.2 | 3.2 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.2 | 3.7 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.2 | 3.9 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.2 | 1.7 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.2 | 0.7 | GO:0036233 | glycine import(GO:0036233) |
| 0.2 | 2.9 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.2 | 2.9 | GO:0071415 | cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.2 | 3.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.2 | 2.9 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.2 | 0.7 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.2 | 1.5 | GO:0035815 | positive regulation of renal sodium excretion(GO:0035815) |
| 0.2 | 4.6 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.2 | 1.5 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.2 | 1.1 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.2 | 0.6 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.2 | 1.9 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.2 | 1.4 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.2 | 3.0 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.2 | 4.1 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.2 | 1.8 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.2 | 1.0 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.2 | 4.5 | GO:0045821 | positive regulation of glycolytic process(GO:0045821) |
| 0.2 | 1.5 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.2 | 0.4 | GO:0043032 | positive regulation of macrophage activation(GO:0043032) |
| 0.2 | 1.5 | GO:0045780 | positive regulation of alkaline phosphatase activity(GO:0010694) positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.2 | 1.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.2 | 1.1 | GO:1900107 | embryonic hindgut morphogenesis(GO:0048619) regulation of nodal signaling pathway(GO:1900107) |
| 0.2 | 4.6 | GO:0007616 | long-term memory(GO:0007616) |
| 0.2 | 2.9 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.2 | 0.5 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.2 | 1.9 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.2 | 1.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.2 | 2.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.2 | 1.2 | GO:0006477 | protein sulfation(GO:0006477) heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 1.7 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.2 | 7.5 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.2 | 1.4 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 1.0 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.4 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 | 1.4 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.1 | 3.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 1.4 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.1 | 3.9 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.1 | 4.5 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 0.6 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.1 | 1.9 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 2.1 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.1 | 2.0 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 1.1 | GO:0036507 | protein demannosylation(GO:0036507) protein alpha-1,2-demannosylation(GO:0036508) endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.1 | 1.9 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 1.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.5 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.3 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.1 | 2.1 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.1 | 3.8 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.1 | 0.4 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.1 | 2.7 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 1.9 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.1 | 1.5 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 1.3 | GO:0090344 | negative regulation of cell aging(GO:0090344) |
| 0.1 | 0.2 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 1.8 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 2.4 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.1 | 0.3 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) response to fluoride(GO:1902617) |
| 0.1 | 2.4 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 4.7 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.1 | 4.3 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 1.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.3 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.1 | 10.3 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 1.3 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.1 | 2.7 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 4.9 | GO:0021795 | cerebral cortex cell migration(GO:0021795) |
| 0.1 | 3.3 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 2.7 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.1 | 2.9 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 1.6 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 1.1 | GO:0042711 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.1 | 1.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 4.5 | GO:0051341 | regulation of oxidoreductase activity(GO:0051341) |
| 0.1 | 0.8 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 5.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 3.7 | GO:0006584 | catecholamine metabolic process(GO:0006584) catechol-containing compound metabolic process(GO:0009712) |
| 0.1 | 0.7 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.1 | 0.6 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.1 | 1.9 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 2.1 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 2.3 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.1 | 0.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 1.1 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 0.6 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 2.1 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.1 | 0.1 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.1 | 7.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.6 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.1 | 1.2 | GO:0006525 | arginine metabolic process(GO:0006525) |
| 0.1 | 1.7 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.1 | 2.0 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.1 | 0.4 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 11.6 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.1 | 6.3 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 4.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 2.3 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.1 | 3.5 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 | 4.0 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 1.9 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.2 | GO:0038042 | dimeric G-protein coupled receptor signaling pathway(GO:0038042) calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 3.5 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 3.1 | GO:0009798 | axis specification(GO:0009798) |
| 0.0 | 2.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 3.0 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 1.1 | GO:0009083 | branched-chain amino acid metabolic process(GO:0009081) branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 4.8 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 4.8 | GO:0044782 | cilium organization(GO:0044782) |
| 0.0 | 0.8 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 1.0 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 1.3 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 1.4 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.1 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 2.7 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 1.5 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.1 | GO:0021590 | hindbrain maturation(GO:0021578) cerebellum maturation(GO:0021590) central nervous system maturation(GO:0021626) cerebellar cortex maturation(GO:0021699) |
| 0.0 | 1.4 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 1.5 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 2.5 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.4 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.6 | GO:0060384 | innervation(GO:0060384) |
| 0.0 | 0.5 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.7 | GO:0003407 | neural retina development(GO:0003407) |
| 0.0 | 0.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.4 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.7 | GO:0071347 | cellular response to interleukin-1(GO:0071347) |
| 0.0 | 0.4 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.4 | 43.2 | GO:0071750 | dimeric IgA immunoglobulin complex(GO:0071750) secretory dimeric IgA immunoglobulin complex(GO:0071752) |
| 3.1 | 18.8 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 2.5 | 7.4 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 2.2 | 6.7 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 1.7 | 83.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 1.3 | 5.4 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 1.3 | 11.6 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 1.2 | 3.5 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 1.0 | 6.3 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.9 | 169.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.7 | 11.8 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.7 | 17.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.7 | 3.9 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.6 | 3.1 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.6 | 3.9 | GO:0043196 | varicosity(GO:0043196) |
| 0.6 | 4.4 | GO:0030891 | VCB complex(GO:0030891) Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.5 | 2.7 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.5 | 4.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.5 | 2.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.5 | 5.0 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.5 | 1.4 | GO:0098536 | deuterosome(GO:0098536) |
| 0.4 | 1.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.4 | 2.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.4 | 5.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.4 | 3.6 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.4 | 4.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.4 | 2.3 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.4 | 30.6 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.3 | 5.8 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.3 | 2.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.3 | 4.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.3 | 1.5 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.3 | 1.4 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.3 | 11.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.3 | 1.9 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.3 | 1.9 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.3 | 7.2 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.3 | 1.8 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.3 | 11.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.3 | 1.0 | GO:0071546 | pi-body(GO:0071546) |
| 0.3 | 18.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 3.4 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.2 | 2.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.2 | 1.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.2 | 14.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 19.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 1.7 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.2 | 1.4 | GO:0031088 | platelet dense granule membrane(GO:0031088) platelet dense granule(GO:0042827) |
| 0.2 | 2.8 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.2 | 6.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.2 | 1.5 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.2 | 3.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.2 | 8.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.2 | 27.0 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.2 | 0.5 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.2 | 10.3 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.2 | 4.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.2 | 0.5 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.2 | 229.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 1.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 2.9 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 1.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.4 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 2.0 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 2.5 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 3.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 1.6 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.1 | 3.5 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 0.8 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 3.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 6.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 4.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 10.9 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.1 | 0.5 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 1.7 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 3.1 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 0.6 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 2.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 5.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 1.0 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 3.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 1.7 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 1.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.6 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.8 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 7.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 7.3 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 2.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 12.2 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 1.5 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 2.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.8 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 3.3 | GO:0000428 | DNA-directed RNA polymerase complex(GO:0000428) |
| 0.0 | 0.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 2.5 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.4 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.5 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.5 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 4.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 5.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.5 | GO:0098858 | actin-based cell projection(GO:0098858) |
| 0.0 | 0.5 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.6 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.8 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.9 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 4.1 | GO:0043025 | neuronal cell body(GO:0043025) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.3 | 28.0 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 5.5 | 21.9 | GO:0032810 | sterol response element binding(GO:0032810) |
| 4.7 | 18.8 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 2.9 | 11.6 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 2.8 | 14.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 2.6 | 608.7 | GO:0003823 | antigen binding(GO:0003823) |
| 2.5 | 15.1 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 2.5 | 7.4 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 2.4 | 9.7 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 2.4 | 12.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 2.4 | 12.0 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 2.0 | 17.8 | GO:0043426 | MRF binding(GO:0043426) |
| 1.9 | 5.7 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 1.8 | 11.0 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 1.7 | 6.8 | GO:0004341 | gluconolactonase activity(GO:0004341) |
| 1.5 | 4.6 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 1.5 | 12.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 1.5 | 5.8 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 1.4 | 5.5 | GO:0017089 | glycolipid transporter activity(GO:0017089) ceramide binding(GO:0097001) |
| 1.4 | 5.5 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 1.3 | 3.9 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 1.2 | 6.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 1.1 | 3.3 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 1.1 | 5.3 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 1.0 | 5.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 1.0 | 9.8 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.9 | 14.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.9 | 2.8 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.9 | 4.5 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.9 | 7.9 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.9 | 3.5 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.8 | 3.1 | GO:0023029 | MHC class Ib protein binding(GO:0023029) |
| 0.8 | 10.9 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.8 | 2.3 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.8 | 11.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.7 | 6.7 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.7 | 4.5 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.7 | 8.8 | GO:0008430 | selenium binding(GO:0008430) |
| 0.7 | 21.8 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.7 | 18.0 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.7 | 2.1 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.7 | 2.0 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.7 | 2.0 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.7 | 2.0 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.6 | 7.1 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.6 | 7.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.6 | 6.3 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.6 | 3.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.6 | 3.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.6 | 6.7 | GO:0004568 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.6 | 3.0 | GO:0004920 | type I interferon receptor activity(GO:0004905) interleukin-10 receptor activity(GO:0004920) |
| 0.6 | 3.6 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) |
| 0.6 | 2.4 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.6 | 4.6 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.6 | 4.4 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.5 | 2.7 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.5 | 3.7 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.5 | 2.6 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.5 | 2.0 | GO:0002046 | opsin binding(GO:0002046) |
| 0.5 | 7.3 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.5 | 4.7 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.5 | 3.7 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.5 | 4.5 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.5 | 1.8 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.5 | 6.8 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.5 | 2.3 | GO:0004471 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) oxaloacetate decarboxylase activity(GO:0008948) |
| 0.4 | 1.7 | GO:0004077 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.4 | 2.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.4 | 2.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.4 | 2.0 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.4 | 5.6 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.4 | 7.6 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.4 | 1.2 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.4 | 1.9 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.4 | 1.1 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.4 | 2.6 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.4 | 3.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.4 | 3.5 | GO:0015645 | fatty acid ligase activity(GO:0015645) |
| 0.4 | 3.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.4 | 37.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.3 | 9.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.3 | 4.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.3 | 4.3 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.3 | 4.6 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.3 | 4.2 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.3 | 2.9 | GO:0089720 | caspase binding(GO:0089720) |
| 0.3 | 1.3 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.3 | 9.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.3 | 1.2 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.3 | 4.6 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.3 | 1.2 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.3 | 1.2 | GO:0047291 | neolactotetraosylceramide alpha-2,3-sialyltransferase activity(GO:0004513) lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.3 | 1.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.3 | 4.0 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.3 | 2.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) heme transporter activity(GO:0015232) |
| 0.3 | 1.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.3 | 1.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.3 | 0.8 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.3 | 1.5 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 2.0 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.2 | 2.2 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.2 | 2.8 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.2 | 1.1 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.2 | 3.8 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.2 | 2.9 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.2 | 6.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 3.9 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.2 | 3.9 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.2 | 3.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 1.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 7.0 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.2 | 2.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 0.8 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 1.0 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.2 | 35.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.2 | 42.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 1.5 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.2 | 7.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 12.3 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.2 | 3.9 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.2 | 9.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.2 | 1.5 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.2 | 11.1 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.2 | 1.5 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.2 | 1.5 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 5.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 8.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 3.9 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.4 | GO:0017153 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.1 | 1.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 1.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 1.8 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.1 | 2.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 2.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 1.8 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 1.4 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 1.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 0.5 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.1 | 0.8 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.1 | 12.8 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 1.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 1.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 0.7 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 5.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.7 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 2.1 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.1 | 2.5 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.1 | 0.6 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 2.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 1.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 2.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.5 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 1.8 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 5.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 2.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.5 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.8 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 5.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 4.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 1.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.3 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.1 | 3.3 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.1 | 2.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 6.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 4.8 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 0.6 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.2 | GO:0004948 | calcitonin receptor activity(GO:0004948) calcitonin receptor binding(GO:0031716) |
| 0.1 | 1.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 1.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 2.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.9 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 3.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 1.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.2 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 3.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.6 | GO:0001163 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 1.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 4.4 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.0 | 0.6 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 1.0 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 2.8 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 2.2 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 1.7 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 2.7 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.1 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 4.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 1.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 4.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 1.8 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.1 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.7 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 1.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 5.9 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 1.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.4 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 29.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.5 | 12.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.5 | 38.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.3 | 17.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.3 | 3.7 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.3 | 12.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.3 | 7.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 13.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 2.1 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.2 | 3.0 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 6.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.2 | 31.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.2 | 9.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 3.9 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 2.2 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.2 | 9.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 3.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 14.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.2 | 8.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.2 | 8.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.2 | 1.9 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 6.8 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 6.7 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 9.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 6.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 4.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 2.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 3.9 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 2.7 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 3.5 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 5.3 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 2.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 2.0 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 4.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 5.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 3.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 2.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 3.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 1.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 1.8 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 4.1 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 9.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.1 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.6 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 4.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 21.0 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 1.3 | 12.8 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 1.2 | 12.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 1.1 | 2.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.8 | 17.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.7 | 11.6 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.7 | 15.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.6 | 27.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.5 | 9.2 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.5 | 15.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.5 | 9.5 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.5 | 8.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.4 | 5.4 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.4 | 6.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.4 | 4.4 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.4 | 18.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.4 | 4.9 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.4 | 4.8 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.3 | 32.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.3 | 5.0 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.3 | 8.0 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.3 | 2.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 6.7 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.2 | 20.0 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.2 | 1.5 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.2 | 6.0 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.2 | 9.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 30.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 2.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.2 | 2.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.2 | 3.0 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 12.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 12.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 1.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 3.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 1.9 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 16.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 10.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 2.8 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.2 | 3.2 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.2 | 13.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 18.5 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 7.2 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 7.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 2.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 4.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 2.0 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 2.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 2.3 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.1 | 4.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 5.1 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 5.9 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.1 | 1.0 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 0.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 0.8 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.2 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 0.8 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.9 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 3.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 2.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 7.4 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 1.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 2.2 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 1.1 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 2.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.0 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.7 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 1.6 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 2.6 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 0.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 3.9 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |