Project
avrg: GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
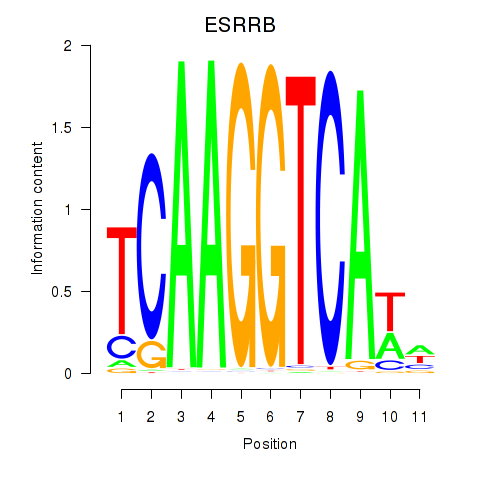
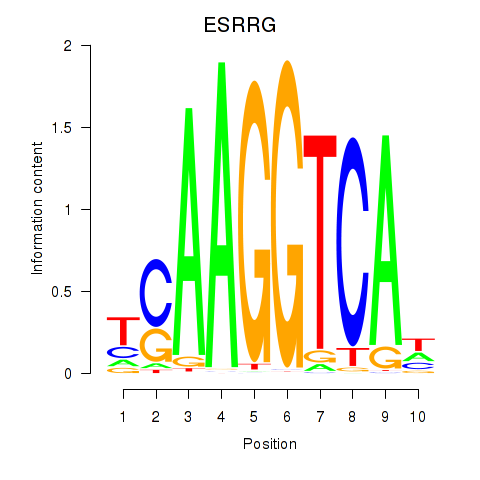
Results for ESRRB_ESRRG
Z-value: 8.75
Transcription factors associated with ESRRB_ESRRG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ESRRB
|
ENSG00000119715.15 | ESRRB |
|
ESRRG
|
ENSG00000196482.18 | ESRRG |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ESRRG | hg38_v1_chr1_-_217076889_217076997 | -0.54 | 6.4e-18 | Click! |
Activity profile of ESRRB_ESRRG motif
Sorted Z-values of ESRRB_ESRRG motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ESRRB_ESRRG
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 32.1 | 128.5 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 20.1 | 60.4 | GO:2000276 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 18.8 | 56.4 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 18.1 | 72.3 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 17.0 | 67.9 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 13.7 | 82.2 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 13.7 | 13.7 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 12.4 | 99.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 12.3 | 49.3 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 12.0 | 48.0 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 12.0 | 35.9 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 11.8 | 106.0 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 11.0 | 33.0 | GO:0002368 | B cell cytokine production(GO:0002368) |
| 10.8 | 204.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 9.5 | 56.7 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 9.3 | 46.5 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 9.3 | 37.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 9.2 | 27.6 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 8.4 | 33.4 | GO:1904045 | cellular response to aldosterone(GO:1904045) |
| 8.3 | 33.1 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 8.0 | 23.9 | GO:0019046 | release from viral latency(GO:0019046) regulation of DNA strand elongation(GO:0060382) |
| 7.7 | 23.2 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 7.7 | 23.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 7.5 | 82.4 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 7.4 | 22.3 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 6.9 | 6.9 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 6.6 | 79.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 6.3 | 19.0 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 5.6 | 67.3 | GO:0042407 | cristae formation(GO:0042407) |
| 5.4 | 27.2 | GO:0021699 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 4.8 | 4.8 | GO:0002339 | B cell selection(GO:0002339) |
| 4.8 | 304.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 4.7 | 88.7 | GO:0075713 | establishment of integrated proviral latency(GO:0075713) |
| 4.4 | 65.5 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 4.2 | 33.7 | GO:0044206 | pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) |
| 3.8 | 11.3 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 3.8 | 33.8 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 3.7 | 37.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 3.5 | 13.9 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 3.5 | 20.8 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 3.4 | 41.4 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 3.4 | 10.3 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 3.4 | 13.6 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 3.3 | 36.3 | GO:0007144 | female meiosis I(GO:0007144) |
| 3.3 | 19.7 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 3.1 | 12.5 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 3.0 | 26.8 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 2.9 | 20.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 2.9 | 26.0 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 2.9 | 31.5 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 2.8 | 13.8 | GO:0030242 | pexophagy(GO:0030242) |
| 2.7 | 8.2 | GO:1903352 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 2.7 | 10.8 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 2.7 | 42.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 2.6 | 13.0 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 2.6 | 13.0 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 2.6 | 12.9 | GO:1903027 | regulation of opsonization(GO:1903027) |
| 2.5 | 10.1 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 2.5 | 60.8 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 2.5 | 25.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 2.5 | 67.2 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 2.4 | 41.6 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 2.4 | 7.3 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 2.4 | 12.1 | GO:1901911 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 2.4 | 9.5 | GO:0003290 | septum secundum development(GO:0003285) atrial septum secundum morphogenesis(GO:0003290) |
| 2.4 | 7.1 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 2.3 | 32.2 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 2.2 | 15.7 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 2.2 | 22.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 2.2 | 13.2 | GO:0097338 | response to clozapine(GO:0097338) |
| 2.2 | 19.7 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 2.2 | 88.2 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 2.0 | 38.4 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 1.8 | 27.7 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 1.8 | 16.5 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 1.8 | 3.6 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 1.8 | 7.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 1.8 | 5.3 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 1.8 | 131.8 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 1.7 | 10.0 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 1.6 | 6.5 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 1.6 | 45.6 | GO:0042026 | protein refolding(GO:0042026) |
| 1.6 | 38.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 1.6 | 15.8 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 1.5 | 20.9 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 1.5 | 23.9 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 1.5 | 31.3 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 1.5 | 7.4 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 1.4 | 25.6 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 1.4 | 7.1 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 1.4 | 12.6 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 1.4 | 18.2 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 1.4 | 39.0 | GO:0033599 | regulation of mammary gland epithelial cell proliferation(GO:0033599) |
| 1.4 | 17.6 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 1.3 | 31.6 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 1.3 | 11.4 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 1.2 | 3.7 | GO:0042323 | negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 1.2 | 6.2 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 1.2 | 12.2 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 1.2 | 13.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 1.1 | 34.1 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 1.1 | 2.2 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 1.1 | 6.3 | GO:0010734 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 1.0 | 13.2 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 1.0 | 1.9 | GO:0034759 | detoxification of copper ion(GO:0010273) regulation of iron ion transport(GO:0034756) regulation of iron ion transmembrane transport(GO:0034759) stress response to copper ion(GO:1990169) |
| 0.9 | 4.7 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.9 | 4.6 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.9 | 13.8 | GO:0061339 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.9 | 7.8 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.8 | 0.8 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.8 | 6.7 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.8 | 2.5 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.8 | 14.1 | GO:0046852 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.8 | 2.5 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.8 | 6.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.7 | 2.9 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.7 | 11.7 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.7 | 5.9 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.7 | 41.3 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.7 | 48.0 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.7 | 26.6 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.7 | 11.1 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.7 | 15.8 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.7 | 8.2 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.7 | 12.7 | GO:0060973 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) cell migration involved in heart development(GO:0060973) |
| 0.6 | 17.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.6 | 17.4 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.6 | 9.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.6 | 13.9 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.6 | 7.5 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.6 | 3.7 | GO:1903243 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of lung blood pressure(GO:0061767) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.6 | 22.3 | GO:0061641 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.6 | 1.8 | GO:1903281 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.6 | 9.7 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.6 | 17.5 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.6 | 9.1 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.6 | 6.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.6 | 2.8 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.5 | 9.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.5 | 3.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.5 | 12.7 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.5 | 3.7 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.5 | 5.7 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.5 | 6.6 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.5 | 7.5 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.5 | 65.5 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.5 | 11.8 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.5 | 14.3 | GO:0046655 | tetrahydrofolate metabolic process(GO:0046653) folic acid metabolic process(GO:0046655) |
| 0.5 | 1.9 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.5 | 4.8 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.5 | 1.9 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.5 | 6.5 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.5 | 5.1 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.4 | 5.2 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.4 | 3.0 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.4 | 2.6 | GO:0060356 | leucine import(GO:0060356) |
| 0.4 | 7.5 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.4 | 9.0 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.4 | 2.0 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.4 | 6.3 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.4 | 1.9 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.4 | 3.0 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.4 | 2.9 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.4 | 81.9 | GO:0006457 | protein folding(GO:0006457) |
| 0.3 | 7.7 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.3 | 11.1 | GO:0035825 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.3 | 33.1 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.3 | 5.5 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.3 | 15.5 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.3 | 27.9 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.3 | 11.0 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.3 | 7.8 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.3 | 13.8 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.3 | 7.2 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.2 | 2.5 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.2 | 5.6 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.2 | 5.0 | GO:0007614 | short-term memory(GO:0007614) |
| 0.2 | 11.9 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.2 | 1.1 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.2 | 0.6 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.2 | 5.8 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.2 | 2.0 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.2 | 2.0 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.2 | 7.3 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.2 | 3.9 | GO:1901032 | negative regulation of response to reactive oxygen species(GO:1901032) negative regulation of hydrogen peroxide-induced cell death(GO:1903206) |
| 0.2 | 2.8 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.2 | 2.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 1.9 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.2 | 2.6 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.2 | 7.3 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 10.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 10.9 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 3.7 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 | 1.9 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.1 | 8.4 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.1 | 3.1 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 1.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 5.7 | GO:0043620 | regulation of DNA-templated transcription in response to stress(GO:0043620) |
| 0.1 | 0.7 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.1 | 1.3 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.1 | 3.0 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 8.9 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.1 | 1.9 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.1 | 5.6 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.1 | 7.7 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.1 | 0.2 | GO:0045656 | negative regulation of monocyte differentiation(GO:0045656) |
| 0.1 | 14.1 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.1 | 6.9 | GO:0050905 | neuromuscular process(GO:0050905) |
| 0.1 | 1.8 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.1 | 10.1 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.1 | 4.3 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 0.6 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 2.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.2 | GO:1903795 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) regulation of intracellular calcium activated chloride channel activity(GO:1902938) regulation of inorganic anion transmembrane transport(GO:1903795) |
| 0.0 | 0.7 | GO:1901798 | positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 1.8 | GO:2001022 | positive regulation of response to DNA damage stimulus(GO:2001022) |
| 0.0 | 1.7 | GO:0060071 | Wnt signaling pathway, planar cell polarity pathway(GO:0060071) |
| 0.0 | 0.4 | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 | 0.3 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.4 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.5 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.0 | 0.2 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.9 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.3 | 225.0 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 15.7 | 94.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 12.3 | 61.3 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 11.8 | 129.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 10.6 | 63.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 10.3 | 82.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 10.3 | 92.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 8.6 | 42.8 | GO:0070470 | plasma membrane respiratory chain complex I(GO:0045272) plasma membrane respiratory chain(GO:0070470) |
| 6.1 | 18.3 | GO:0016939 | kinesin II complex(GO:0016939) |
| 6.1 | 24.4 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 6.0 | 65.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 5.5 | 49.3 | GO:0097452 | GAIT complex(GO:0097452) |
| 4.8 | 24.2 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 4.5 | 353.3 | GO:0098800 | inner mitochondrial membrane protein complex(GO:0098800) |
| 4.4 | 39.8 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 4.1 | 41.0 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 4.0 | 27.7 | GO:0031415 | NatA complex(GO:0031415) |
| 3.8 | 151.8 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 3.4 | 23.9 | GO:0032021 | NELF complex(GO:0032021) |
| 3.0 | 15.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 2.9 | 35.2 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 2.9 | 5.9 | GO:0060203 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 2.8 | 36.8 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 2.8 | 33.8 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 2.8 | 47.1 | GO:0070469 | respiratory chain(GO:0070469) |
| 2.7 | 141.9 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 2.5 | 7.4 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 2.4 | 48.5 | GO:0033270 | paranode region of axon(GO:0033270) |
| 2.4 | 16.6 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 2.4 | 7.1 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 2.3 | 13.8 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 2.2 | 15.7 | GO:1990635 | proximal dendrite(GO:1990635) |
| 1.8 | 25.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 1.7 | 24.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 1.7 | 442.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 1.7 | 31.5 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 1.6 | 28.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 1.5 | 5.9 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 1.5 | 30.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 1.4 | 11.4 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 1.3 | 8.8 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 1.3 | 7.5 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 1.0 | 14.4 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 1.0 | 83.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 1.0 | 2.0 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 1.0 | 26.3 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.9 | 40.9 | GO:0031430 | M band(GO:0031430) |
| 0.8 | 8.4 | GO:0097433 | dense body(GO:0097433) |
| 0.8 | 25.1 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.8 | 7.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.8 | 6.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.7 | 4.5 | GO:0005879 | axonemal microtubule(GO:0005879) asymmetric synapse(GO:0032279) |
| 0.7 | 20.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.7 | 20.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.6 | 17.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.6 | 5.0 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.6 | 7.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.6 | 2.8 | GO:0061202 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.5 | 6.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.5 | 32.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.4 | 6.7 | GO:0046930 | pore complex(GO:0046930) |
| 0.4 | 13.7 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.4 | 15.8 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.4 | 6.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.4 | 3.7 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.4 | 14.4 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.4 | 17.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.3 | 86.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.3 | 51.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.3 | 93.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.3 | 6.6 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.3 | 10.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.3 | 25.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.3 | 3.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.3 | 1.8 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.3 | 6.1 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.2 | 6.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.2 | 6.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 26.5 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.2 | 85.1 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.2 | 3.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.2 | 28.0 | GO:0000922 | spindle pole(GO:0000922) |
| 0.2 | 1.9 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.2 | 28.4 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.1 | 10.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.8 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 7.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 21.9 | GO:0031968 | organelle outer membrane(GO:0031968) |
| 0.1 | 1.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 25.1 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 2.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 39.4 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 3.1 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 4.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 1.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 12.4 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.1 | 5.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 1.9 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 0.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 9.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 5.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) zonula adherens(GO:0005915) |
| 0.0 | 1.3 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.4 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 24.8 | 99.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 17.0 | 67.9 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 16.4 | 49.3 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 15.3 | 61.3 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 11.9 | 95.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 10.3 | 82.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 9.9 | 79.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 8.8 | 43.9 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 8.4 | 91.9 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 8.3 | 24.8 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 7.7 | 38.4 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 7.5 | 217.0 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 6.4 | 25.5 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 5.8 | 46.6 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 5.5 | 33.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 5.3 | 281.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 5.0 | 15.0 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 5.0 | 89.8 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 4.8 | 19.0 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 4.5 | 22.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 4.2 | 33.8 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 4.2 | 20.8 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 3.9 | 31.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 3.7 | 48.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 3.7 | 11.0 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 3.6 | 54.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 3.6 | 14.3 | GO:0004487 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 3.5 | 76.9 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 3.4 | 10.3 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 3.4 | 20.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 3.4 | 10.1 | GO:0009041 | uridylate kinase activity(GO:0009041) nucleoside phosphate kinase activity(GO:0050145) |
| 3.3 | 22.9 | GO:0004459 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 3.3 | 39.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 3.1 | 9.4 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 3.0 | 39.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 2.8 | 33.4 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 2.7 | 8.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 2.5 | 10.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 2.5 | 20.2 | GO:0015288 | porin activity(GO:0015288) |
| 2.5 | 67.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 2.4 | 7.3 | GO:0031403 | lithium ion binding(GO:0031403) |
| 2.4 | 12.1 | GO:0052842 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 2.4 | 11.8 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 2.3 | 11.5 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 2.2 | 10.8 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 2.1 | 39.9 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 2.0 | 27.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 2.0 | 21.7 | GO:0033691 | sialic acid binding(GO:0033691) |
| 1.9 | 7.5 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 1.7 | 12.2 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 1.7 | 211.4 | GO:0051087 | chaperone binding(GO:0051087) |
| 1.7 | 5.1 | GO:0031753 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 1.6 | 4.8 | GO:0048030 | disaccharide binding(GO:0048030) |
| 1.5 | 13.7 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 1.5 | 98.9 | GO:0009055 | electron carrier activity(GO:0009055) |
| 1.4 | 7.2 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 1.4 | 11.4 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 1.4 | 5.6 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 1.4 | 8.4 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 1.4 | 9.5 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 1.4 | 34.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 1.3 | 66.0 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 1.3 | 4.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 1.3 | 5.3 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 1.2 | 12.3 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 1.2 | 3.6 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 1.1 | 6.9 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 1.1 | 21.4 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.9 | 5.5 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.9 | 9.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.9 | 10.0 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.9 | 13.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.8 | 11.3 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.7 | 3.7 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.7 | 9.5 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.7 | 16.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.7 | 12.8 | GO:0031005 | filamin binding(GO:0031005) |
| 0.7 | 18.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.6 | 13.4 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.6 | 5.7 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.6 | 8.8 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.6 | 135.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.6 | 17.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.6 | 3.7 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.6 | 2.5 | GO:0033265 | choline binding(GO:0033265) |
| 0.6 | 33.1 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.6 | 5.9 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.6 | 12.4 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.6 | 4.6 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.5 | 3.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.5 | 61.5 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.5 | 9.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.5 | 11.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.5 | 12.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.5 | 2.9 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.5 | 14.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.5 | 31.6 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.5 | 15.8 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.5 | 9.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.5 | 6.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.4 | 0.8 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.4 | 70.2 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.4 | 6.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.4 | 12.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.4 | 5.7 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.4 | 3.0 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.4 | 1.8 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.4 | 2.5 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.3 | 23.3 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.3 | 10.0 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.3 | 14.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.3 | 2.8 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.3 | 6.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.3 | 7.8 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.3 | 1.5 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.3 | 7.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.3 | 1.2 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.3 | 1.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.3 | 15.7 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.3 | 2.4 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.2 | 1.7 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.2 | 9.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.2 | 36.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 13.1 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.2 | 7.2 | GO:0043531 | ADP binding(GO:0043531) |
| 0.2 | 29.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.2 | 6.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.2 | 0.6 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.2 | 25.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.2 | 6.2 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.2 | 13.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 4.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 2.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.2 | 19.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 7.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.2 | 21.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.2 | 6.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 33.4 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 2.0 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.1 | 9.9 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 8.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 4.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 29.6 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 10.6 | GO:0016829 | lyase activity(GO:0016829) |
| 0.1 | 3.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 5.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 15.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 3.9 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 6.7 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 2.9 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 0.7 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 3.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 4.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 5.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 2.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 5.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 3.8 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 1.9 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 2.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 2.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 48.0 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 3.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 3.7 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 0.3 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.9 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 36.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 1.8 | 256.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 1.1 | 34.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 1.1 | 24.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 1.0 | 19.7 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.8 | 89.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.6 | 12.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.5 | 22.3 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.5 | 4.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.5 | 39.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.5 | 32.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.5 | 27.0 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.5 | 13.8 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.4 | 46.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.4 | 29.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.4 | 7.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.4 | 20.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.4 | 7.0 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.3 | 8.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.3 | 5.7 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.3 | 29.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.3 | 17.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.3 | 18.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 13.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 10.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 4.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 6.3 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 6.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 5.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 2.0 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 5.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.5 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 7.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 3.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 11.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.2 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.2 | 781.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 8.1 | 88.7 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 5.1 | 76.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 4.3 | 283.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 4.0 | 220.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 2.2 | 24.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 2.0 | 31.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 1.9 | 64.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 1.9 | 34.9 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 1.8 | 27.6 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 1.7 | 33.2 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 1.4 | 32.6 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 1.3 | 38.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 1.3 | 36.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 1.1 | 31.9 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 1.0 | 22.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.9 | 25.0 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.8 | 11.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.8 | 25.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.7 | 15.8 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.7 | 10.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.7 | 18.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.6 | 23.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.6 | 11.9 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.6 | 12.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.6 | 11.5 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.6 | 21.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.6 | 147.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.5 | 16.7 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.5 | 6.2 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.4 | 9.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.3 | 7.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.3 | 3.7 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.3 | 30.3 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.3 | 7.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.3 | 6.7 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 5.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.2 | 7.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 5.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 8.0 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.2 | 3.7 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 5.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 31.8 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.2 | 3.9 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.2 | 2.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 5.7 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 3.7 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 4.9 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 4.8 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 4.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 5.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 3.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 2.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 5.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.7 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 3.0 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 13.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 2.7 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 3.1 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 5.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 2.8 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |